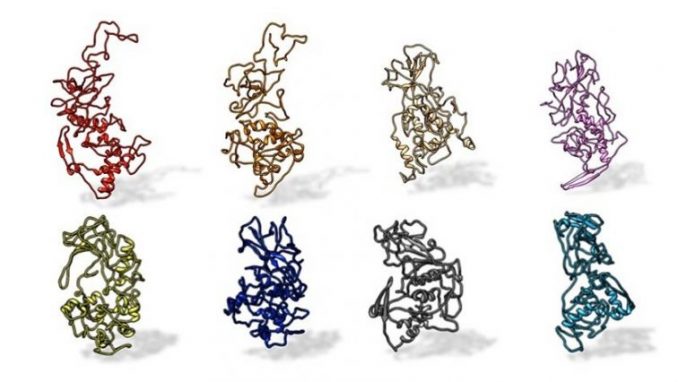
A Penn State research study group discovered that the N protein on SARS-CoV-2 is saved throughout all SARS-related pandemic coronaviruses (top, from left: SARS-CoV-2, civet, SARS-CoV, MERS). The protein varies from other coronaviruses, such as those that trigger the acute rhinitis (bottom, from left: OC43, HKU1, NL63, and 229E). Credit: Kelly Lab/Penn State
The structural discovery might have ramifications as a healing target in all SARS-CoV-2 variations.
A small protein of SARS-CoV-2, the coronavirus that triggers COVID-19, might have huge ramifications for future treatments, according to a group of Penn State scientists.
Using an unique toolkit of methods, the researchers revealed the very first complete structure of the Nucleocapsid (N) protein and found how antibodies from COVID-19 clients connect with that protein. They likewise identified that the structure appears comparable throughout numerous coronaviruses, consisting of current COVID-19 variations — making it a perfect target for innovative treatments and vaccines. They reported their lead to Nanoscale.
“We discovered new features about the N protein structure that could have large implications in antibody testing and the long-term effects of all SARS-related pandemic viruses,” stated Deb Kelly, teacher of biomedical engineering (BME), Huck Chair in Molecular Biophysics and director of the Penn State Center for Structural Oncology, who led the research study. “Since it appears that the N protein is conserved across the variants of SARS-CoV-2 and SARS-CoV-1, therapeutics designed to target the N protein could potentially help knock out the harsher or lasting symptoms some people experience.”
Most of the diagnostic tests and readily available vaccines for COVID-19 were created based upon a bigger SARS-CoV-2 protein — the Spike protein — where the infection connects to healthy cells to start the intrusion procedure.
The Pfizer/BioNTech and Moderna vaccines were created to assist receivers produce antibodies that secure versus the Spike protein. However, Kelly stated, the Spike protein can quickly alter, leading to the variations that have actually emerged in the United Kingdom, South Africa, Brazil, and throughout the United States.
Unlike the external Spike protein, the N protein is enclosed in the infection, secured from ecological pressures that trigger the Spike protein to alter. In the blood, nevertheless, the N protein drifts easily after it is launched from contaminated cells. The free-roaming protein triggers a strong immune action, causing the production of protective antibodies. Most antibody-testing sets search for the N protein to identify if an individual was formerly contaminated with the infection — rather than diagnostic tests that search for the Spike protein to identify if an individual is presently contaminated.
“Everyone is looking at the Spike protein, and there are fewer studies being performed on the N protein,” stated Michael Casasanta, very first author on the paper and a postdoctoral fellow in the Kelly lab. “There was this gap. We saw an opportunity — we had the ideas and the resources to see what the N protein looks like.”
Initially, the scientists took a look at the N protein series from people, along with various animals believed to be possible sources of the pandemic, such as bats, civets, and pangolins. They all looked comparable however clearly various, according to Casasanta.
“The sequences can predict the structure of each of these N proteins, but you can’t get all the information from a prediction — you need to see the actual 3D structure,” Casasanta stated. “We converged the technology to see a new thing in a new way.”
The scientists utilized an electron microscopic lense to image both the N protein and the website on the N protein where antibodies bind, utilizing serum from COVID-19 clients, and established a 3D computer system design of the structure. They discovered that the antibody binding website stayed the very same throughout every sample, making it a possible target to deal with individuals with any of the recognized COVID-19 variations.
“If a therapeutic can be designed to target the N protein binding site, it might help reduce the inflammation and other lasting immune responses to COVID-19, especially in COVID long haulers,” Kelly stated, describing individuals who experience COVID-19 signs for 6 weeks or longer.
The group obtained cleansed N proteins, indicating the samples just consisted of N proteins, from RayBiotech Life and used them to microchips established in collaboration with Protochips Inc. The microchips are made from silicon nitride, rather than a more standard permeable carbon, and they consist of thin wells with unique finishings that draw in the N proteins to their surface area. Once ready, the samples were flash frozen and taken a look at through cryo-electron microscopy.
Kelly credited her group’s distinct mix of microchips, thinner ice samples and Penn State’s innovative electron microscopic lens equipped with modern detectors, tailored from the business Direct Electron, for providing the highest-resolution visualization of low-weight particles from SARS-CoV-2 up until now.
“The technology combined resulted in a unique finding,” Kelly stated. “Before, it resembled attempting to take a look at something frozen in the middle of the lake. Now, we’re taking a look at it through an ice. We can see smaller sized entities with much more information and greater precision.”
Reference: “Microchip-based structure determination of low-molecular weight proteins using cryo-electron microscopy” by Michael A. Casasanta, G. M. Jonaid, Liam Kaylor, William Y. Luqiu, Maria J. Solares, Mariah L. Schroen, William J. Dearnaley, Jarad Wilson, Madeline J. Dukes and Deborah F. Kelly, 1 April 2021, Nanoscale.
DOI: 10.1039/D1NR00388G
Casasanta and Kelly are both likewise associated with Penn State’s Materials Research Institute (MRI). Co-authors consist of G.M. Jonaid, BME and Bioinformatics and Genomics Graduate Program in Penn State’s Huck Institutes of the Life Sciences; Liam Kaylor and Maria J. Solares, BME and Molecular, Cellular, and Integrative Biosciences Graduate Program in the Huck Institutes of the Life Sciences; William Y. Luqiu, MRI and Department of Electrical and Computer Engineering at Duke University; Mariah Schroen, MRI; William J. Dearnaley, BME and MRI; Jared Wilson, RayBiotech Life; and Madeline J. Dukes, Protochips Inc.
The National Cancer Institute of the National Institutes of Health and the Center for Structural Oncology in the Huck Institutes of the Life Sciences at Penn State moneyed this work.