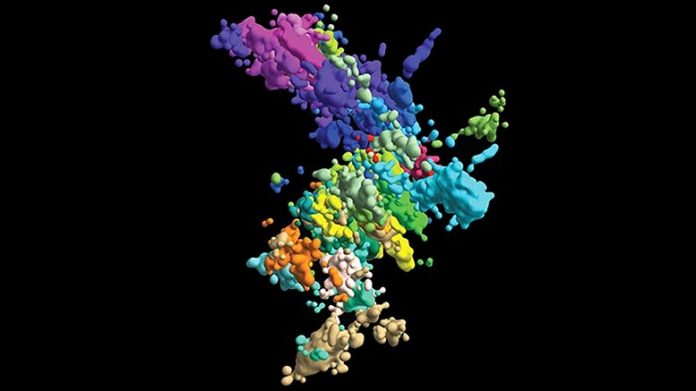
This various colored picture of chromatin was produced utilizing multiplexed fluorescence in situ hybridization and super-resolution microscopy. Credit: Xiaowei Zhuang laboratory
High-resolution, 3D pictures of human chromosomes in single cells expose how DNA structure may affect its function.
In high school books, human chromosomes are envisioned as wonky Xs like 2 hotdogs jammed together. But those images are far from precise. “For 90 percent of the time,” stated Jun-Han Su, “chromosomes don’t exist like that.”
Last year, prior to Su finished with his Ph.D., he and 3 existing Ph.D. prospects in the Graduate School of Arts and Sciences — Pu Zheng, Seon Kinrot and Bogdan Bintu — recorded high-resolution 3D pictures of human chromosomes, the complex homes for our DNA. Now, those images might offer adequate proof to alter those Xs into more complex however much more precise signs to not just teach the next generation of researchers however assist the existing generation decipher secrets about how chromosome structure affects function.
All living things, human beings consisted of, need to develop brand-new cells to change those too old and damaged to operate. To do that, cells divide and reproduce their DNA, which is covered into labyrinthine libraries inside chromatin, the things inside chromosomes. Extended in a straight line, DNA in a single cell can reach 6 feet, all of which gets covered into tight, intricate structures in a cell nucleus. Just one error copying or re-winding that hereditary product might trigger genes to alter or malfunction.
Zooming in close adequate to see chromatin structure is hard. But taking a look at both structure and function is harder still. Now, in a paper released in August in Cell, Zhuang and her group report a brand-new technique to image the structure and habits of chromatin together, linking the dots to figure out how one affects the other to preserve appropriate function or trigger illness.
“It’s quite important to determine the 3D organization,” stated Zhuang, the David B. Arnold, Jr. Professor of Science, “to understand the molecular mechanisms underlying the organization and to also understand how this organization regulates genome function.”
With their brand-new high-resolution 3D imaging technique, the group began to construct a chromosomal map from both wide-lens pictures of all 46 chromosomes and close-ups of one area of one chromosome. To image something that’s still too little to image, they recorded linked dots (“genomic loci”) along each DNA chain. By linking a great deal of dots, they might form a detailed photo of the chromatin structure.
But there was a snag. Previously, Zhuang stated, the variety of dots they might image and determine was restricted by the variety of colors they might image together: 3. Three dots can’t make a detailed photo.
So, Zhuang and her group created a consecutive technique: Image 3 various loci, satiate the signal, and after that image another 3 in quick succession. With that strategy, each dot gets 2 recognizing marks: color and image round.
“Now we actually have 60 loci simultaneously imaged and localized and, importantly, identified,” stated Zhuang.
Still, to cover the entire genome, they required more — thousands — so they relied on a language that’s currently utilized to arrange and save substantial quantities of info: binary. By inscribing binary barcodes on various chromatin loci, they might image much more loci and decipher their identities later on. For example, a particle imaged in round one however not round 2 gets a barcode beginning with “10.” With 20-bit barcodes, the group might distinguish 2,000 particles in simply 20 rounds of imaging. “In this combinatorial way, we can increase the number of molecules that are imaged and identified much more rapidly,” stated Zhuang.
With this strategy, the group imaged about 2,000 chromatin loci per cell, a more than ten-fold boost from their previous work and enough to form a high-resolution picture of what the structure of chromosomes appears like in its native environment. But they didn’t stop there: They likewise imaged transcription activity (when RNA reproduces hereditary product from DNA) and nuclear structures like nuclear speckles and nucleoli.
With their 3D Google Maps of the genome, they might begin to evaluate how the structure moves gradually and how those territorial motions assist or harm cellular division and duplication.
Researchers currently understand chromatin is gotten into various locations and domains (like deserts versus cities). But what those surfaces appear like in various cell types and how they operate is still unidentified. With their high-resolution images, Zhuang and group identified that locations with great deals of genes (“gene-rich”) tend to flock to comparable locations on any chromosome. But locations with couple of genes (“gene-poor”) just come together if they share the exact same chromosome. One theory is that gene-rich locations, which are active websites for gene transcription, come together like a factory to make it possible for more effective production.
While more research study is required prior to verifying this theory, something is now particular: regional chromatin environment affects transcription activity. Structure does affect function. The group likewise found that no 2 chromosomes look the exact same, even in cells that are otherwise similar. To find what each chromosome appears like in every cell in the body will take much more work than one laboratory can handle alone.
“It’s not going to be possible to build just on our work,” Zhuang stated. “We need to build on many, many labs’ work in order to have a comprehensive understanding.”
Reference: “Genome-Scale Imaging of the 3D Organization and Transcriptional Activity of Chromatin” by Jun-Han Su, Pu Zheng, Seon S. Kinrot, Bogdan Bintu and Xiaowei Zhuang, 20 August 2020, Cell.
DOI: 10.1016/j.cell.2020.07.032