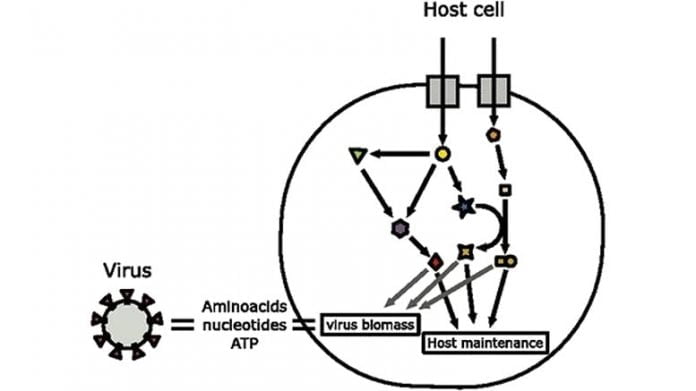
Schematic representation of the incorporated host-virus metabolic modelling technique utilized in the post. The biomass structure of SARS-CoV-2 is approximated based upon genomic and structural details and after that ingrained in the metabolic network design of the host cell. This design is then utilized to forecast the metabolic fluxes supporting infection production and impacts of perturbations. Credit: University of Warwick
- The covid-19 infection, like all infections counts on their host for recreation
- How SARS-CoV-2 records stoichiometric amino and nucleic acids in a human lung cell has actually been found by utilizing a computational design by researchers from the University of Warwick
- By understanding host-based metabolic perturbations hindering SARS-CoV-2, a quick, experimentally testable generation of drug forecasts can be made
A computational design of a human lung cell has actually been utilized to comprehend how SARS-CoV-2 makes use of human host cell metabolic process to recreate by scientists at the University of Warwick. This research study assists comprehend how the infection utilizes the host to endure, and make it possible for drug forecasts for dealing with the infection to be made.
Viruses depend on their host to endure, a vital action of lifecycle is the synthesis of the infection particles within the host cell, for that reason comprehending this procedure is crucial to discovering methods to avoid the infection from enduring.
Using a computer system design of a human lung cell metabolic process, researchers from the School of Life Sciences at the University of Warwick have actually caught the stoichiometric amino and nucleic acid requirements of SARS-CoV-2, the infection that triggers Covid-19. Publishing their lead to the paper, ‘Inhibiting the reproduction of SARS-CoV-2 through perturbations in human lung cell metabolic network’, in the journal Life Science Alliance.
Their design has actually determined host-based metabolic perturbations hindering SARS-CoV-2 recreation, highlighting responses in the main metabolic process, in addition to amino acid and nucleotide biosynthesis paths. In reality, scientists discovered that just few of these metabolic perturbations have the ability to selectively prevent infection recreation.
Researchers have actually likewise kept in mind that a few of the catalysing enzymes of such responses have actually shown interactions with existing drugs, which can be utilized for speculative screening of the provided forecasts utilizing gene knockouts and RNA-disturbance strategies.
Professor Orkun Soyer, from the School of Life Sciences at the University of Warwick remarks:
“We have actually produced a stoichiometric biomass function for the COVID-19-triggering SARS-CoV-2 infection and integrated this into a human lung cell genome scale metabolic design.
“We then predicted reaction perturbations that can inhibit SARS-CoV-2 reproduction in general or selectively, without inhibiting the host metabolic maintenance. The predicted reactions primarily fall onto glycolysis and oxidative phosphorylation pathways, and their connections to amino acid biosynthesis pathways.”
Dr. Hadrien Delattre, from the School of Life Sciences at the University of Warwick includes:
“Together, these outcomes highlight the possibility of targeting host metabolic process for inhibition of SARS-CoV-2 recreation in human cells in basic and in human lung cells particularly.
He included, “More research needs to be carried out to explore SARS-CoV-2 infected cells and their metabolism, however the model developed here by the researchers can be used as a starting point for testing out specific drug predictions.”
Reference: “Inhibiting the reproduction of SARS-CoV-2 through perturbations in human lung cell metabolic network” by Hadrien Delattre, Kalesh Sasidharan and Orkun S Soyer, 24 November 2020, Life Science Alliance.
DOI: 10.26508/lsa.202000869