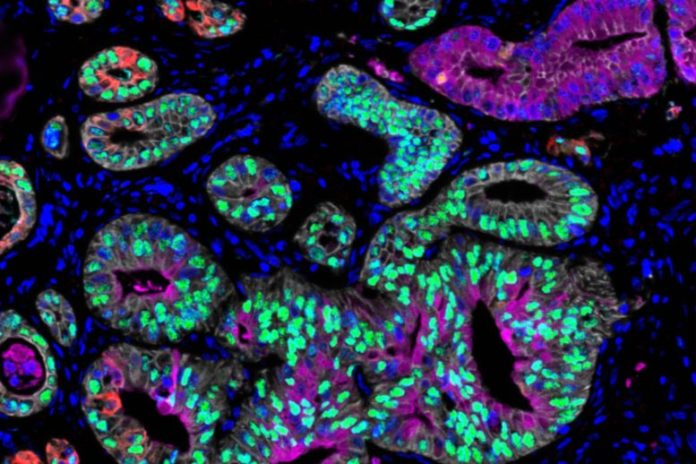
MIT scientists discovered that growth cells from pancreatic cancer clients can exist in 3 states: classical (stained purple and green), basal (stained red), and intermediate (overlapping purple, green and red). Credit: Hannah Williams
Study results likewise reveal that pancreatic growth cells can be pushed into a more prone state by altering their environment.
Over the previous couple of years, researchers have actually made excellent strides in comprehending the hereditary anomalies that can drive cancer. For some kinds of cancer, these discoveries have actually resulted in the advancement of drugs that target particular anomalies.
However, there are still lots of kinds of cancer for which no such targeted treatments are readily available. A group of scientists from MIT, Dana Farber Cancer Institute, and other organizations is now taking a look at whether another cell quality– RNA expression patterns– affects drug actions and can be utilized to determine treatments a growth may be prone to.
In a brand-new research study of pancreatic cancer cells, the scientists determined 3 prototypical RNA-expression states and exposed distinctions in their vulnerability to a range of cancer drugs. They likewise found that changing the growth microenvironment can drive growth cells from one state to another, possibly providing a method to make them more prone to a specific drug.
“What we show in this paper is that cancer cell state is plastic in response to the microenvironment and has a dramatic impact on drug sensitivity. This opens new frontiers for thinking about drug development and how to select drugs for individual patients,” states Alex Shalek, a core member of the Institute for Medical Engineering and Science (IMES) at MIT, an associate teacher of chemistry, and an extramural member of MIT’s Koch Institute for Integrative CancerResearch He is likewise a member of the Ragon Institute of MGH, MIT, and Harvard and an institute member of the Broad Institute.
Shalek and Brian Wolpin, an associate teacher of medication at Harvard Medical School and Dana-Farber Cancer Institute; William Hahn, a teacher of medication at Harvard Medical School and Dana-Farber; and Andrew Aguirre, an assistant teacher of medication at Harvard Medical School and Dana-Farber; are the senior authors of the research study, which was released on December 9, 2021, in Cell The paper’s lead authors are Srivatsan Raghavan, a trainer in medication at Harvard Medical School and Dana-Farber; Peter Winter, an MIT postdoc; Andrew Navia, an MIT college student; and Hannah Williams, a research study fellow in medication at Harvard Medical School and Dana-Farber
Cell states
Sequencing a cell’s genome can expose cancer-linked anomalies, however recognizing these anomalies does not constantly supply details that can be acted on to deal with a specific growth. To create extra information that might be utilized to assist pick more targeted treatments, Shalek and other scientists have actually relied on single-cell RNA-sequencing, which exposes the genes that are being revealed by each cell at a minute in time.
“There are plenty of situations where the genetics are incredibly important, where you can develop these very precise drugs that target mutations or translocations,” Navia states. “But in many instances mutations alone don’t give you an effective way to target cancer cells relative to healthy ones.”
In this research study, the scientists evaluated cells from pancreatic ductal adenocarcinoma (PDAC). There are extremely couple of targeted drugs readily available to deal with pancreatic growths, so most clients get chemotherapy drugs that might work at first however typically quit working as growths end up being resistant. Using single-cell RNA-sequencing, the scientists evaluated about 25 metastatic growth samples from pancreatic cancer clients.
Previous analyses of pancreatic growth cell RNA have actually exposed 2 broad classifications of cell states: basal-like, which is a more aggressive state, and classical. In the brand-new research study, the scientists determined a 3rd state that seems an intermediate in between those 2. Cancer cells might travel through this state as they shift from classical to basal-like, the scientists state.
The scientists likewise discovered that the environment in which cancer cells are grown plays a crucial function in identifying their state. In this research study, they grew matched “organoids,” or small cancer aggregates from each client’s biopsy. Such organoids are typically utilized in accuracy medication pipelines to design growths from specific clients, to assist determine drugs that may be beneficial for those people.
When comparing each in vivo single-cell profile to the matched ex vivo organoid design, the scientists discovered that the organoids typically exist in a various RNA state than cancer cells analyzed straight from the exact same client. “We see the exact same DNA anomalies in the initial growth and its design, however when we begin to analyze what they appear like at the RNA level, we discover that they’re extremely, extremely various,” Shalek states.
That recommends that the state of a growth can be affected by the conditions in which it’s grown instead of its genes alone, he states. The scientists likewise discovered that they might drive cancer cells, even long-established cell line designs, to change in between various states by altering their development conditions. Treating cells with TGF-beta, for instance, drives them to a more aggressive, basal-like state, while taking TGF-beta away leads the cells to go back to the classical state in a meal.
Cells in each of those states depend upon various cell-signaling paths to endure, so understanding the cell state is important to choosing the best sort of drug to deal with a specific growth, the scientists state.
“When we started looking at drug sensitivity, it became very clear that the same model pushed into a different state would respond very differently to a drug,” Navia states. “These state-specific sensitivities become critical as we think about selecting drugs and avoiding resistance. If you don’t know the right state, you could pick the entirely wrong compound and try to target the wrong pathways. If you don’t consider plasticity, the cancer may only respond temporarily until its cells change state.”
Targeted treatment
The findings recommend that more examining the interaction of genes, cell state, and the growth microenvironment might assist scientists to establish brand-new drugs that would efficiently target specific clients’ growths.
“We’re not erasing decades of understanding cancer as a genetic disease, but we’re certainly saying that we need to much better understand the intersection between genetics and state,” Winter states. “Cell state absolutely has ties to the underlying sensitivity of certain models, and therefore patients and to specific drugs.”
The discovery that cancer cells can be driven from one state to another by customizing the signals in their microenvironment raises the possibility of locking cancer cells into a specific state in a foreseeable method by therapeutically changing the growth microenvironment, and after that offering a different drug to target that locked state and improve treatment effectiveness.
With their coworkers at Dana-Farber, the MIT group is now running much bigger drug screens to determine how each drug impacts pancreatic cancer cells in various states. They are likewise studying other kinds of cancer to identify if those cancer cells are likewise able to shift in between various states in action to modifications in their microenvironment.
Reference: “Microenvironment drives cell state, plasticity, and drug response in pancreatic cancer” by Srivatsan Raghavan, Peter S. Winter, Andrew W. Navia, Hannah L. Williams, Alan DenAdel, Kristen E. Lowder, Jennyfer Galvez-Reyes, Radha L. Kalekar, Nolawit Mulugeta, Kevin S. Kapner, Manisha S. Raghavan, Ashir A. Borah, Nuo Liu, Sara A. Väyrynen, Andressa Dias Costa, Raymond W.S. Ng, Junning Wang, Emma K. Hill, Dorisanne Y. Ragon, Lauren K. Brais, Alex M. Jaeger, Liam F. Spurr, Yvonne Y. Li, Andrew D. Cherniack, Matthew A. Booker, Elizabeth F. Cohen, Michael Y. Tolstorukov, Isaac Wakiro, Asaf Rotem, Bruce E. Johnson, James M. McFarland, Ewa T. Sicinska, Tyler E. Jacks, Ryan J. Sullivan, Geoffrey I. Shapiro, Thomas E. Clancy, Kimberly Perez, Douglas A. Rubinson, Kimmie Ng, James M. Cleary, Lorin Crawford, Scott R. Manalis, Jonathan A. Nowak, Brian M. Wolpin, William C. Hahn, Andrew J. Aguirre and Alex K. Shalek, 9 December 2021, Cell.
DOI: 10.1016/ j.cell.202111017
The research study was moneyed, in part, by the National Institutes of Health, the Koch Institute and Dana-Farber/Harvard Cancer Center Bridge Project, the Ludwig Center for Molecular Oncology at MIT, the Beckman Young Investigator Program, a Sloan Fellowship in Chemistry, and the Pew-Stewart Scholars Program for Cancer Research.