This scanning electron microscopic lense image reveals SARS-CoV-2 (round gold items) emerging from the surface area of cells cultured in the laboratory. Credit: NIAID-RML
A CRISPR-based test established at MIT and the Broad Institute can discover almost as numerous cases as the requirement Covid-19 diagnostic.
Since the start of the Covid-19 pandemic, scientists at MIT and the Broad Institute of MIT and Harvard, in addition to their partners at the University of Washington, Fred Hutchinson Cancer Research Center, Brigham and Women’s Hospital, and the Ragon Institute, have actually been dealing with a CRISPR-based diagnostic for Covid-19 that can produce lead to 30 minutes to an hour, with comparable precision as the basic PCR diagnostics now utilized.
The brand-new test, referred to as STOPCovid, is still in the research study phase however, in concept, might be made inexpensively enough that individuals might evaluate themselves every day. In a research study appearing today in the New England Journal of Medicine, the scientists revealed that on a set of client samples, their test discovered 93 percent of the favorable cases as identified by PCR tests for Covid-19.
“We need rapid testing to become part of the fabric of this situation so that people can test themselves every day, which will slow down outbreak,” states Omar Abudayyeh, an MIT McGovern Fellow dealing with the diagnostic.
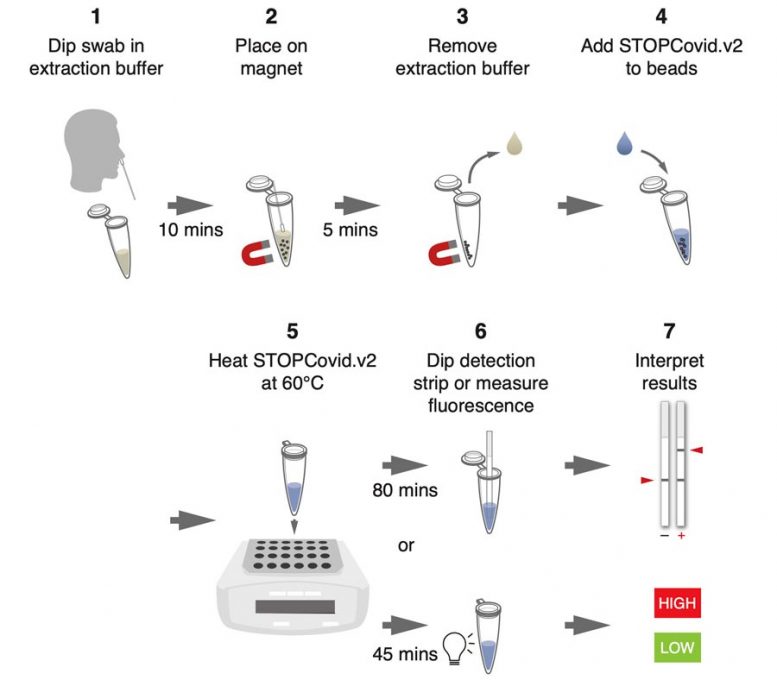
: A nasal swab is dipped in an extraction buffer including a lysis buffer and magnetic beads. After 10 minutes at space temperature level, the sample is put on a magnet and the extraction buffer is aspirated. The STOPCovid response is contributed to the beads, and the sample is heated up to 60 C. For lateral circulation readout, after 80 minutes, detection strips are dipped into the response. For fluorescence readout, after 45 minutes, the response can be analyzed utilizing a fluorescence reader. Credit: Courtesy of the scientists
Abudayyah is among the senior authors of the research study, in addition to Jonathan Gootenberg, a McGovern Fellow, and Feng Zhang, a core member of the Broad Institute, private investigator at the MIT McGovern Institute and Howard Hughes Medical Institute, and the James and Patricia Poitras ’63 Professor of Neuroscience at MIT. The very first authors of the paper are MIT biological engineering college student Julia Joung and Alim Ladha in the Zhang laboratory.
A structured test
Zhang’s lab started working together with the Abudayyeh and Gootenberg lab to deal with the Covid-19 diagnostic right after the SARS-CoV-2 break out started. They concentrated on making an assay, called STOPCovid, that was basic to perform and did not need any customized lab devices. Such a test, they hoped, would be open to future usage in point-of-care settings, such as medical professionals’ workplaces, drug stores, nursing houses, and schools.
“We developed STOPCovid so that everything could be done in a single step,” Joung states. “A single step means the test can be potentially performed by nonexperts outside of laboratory settings.”
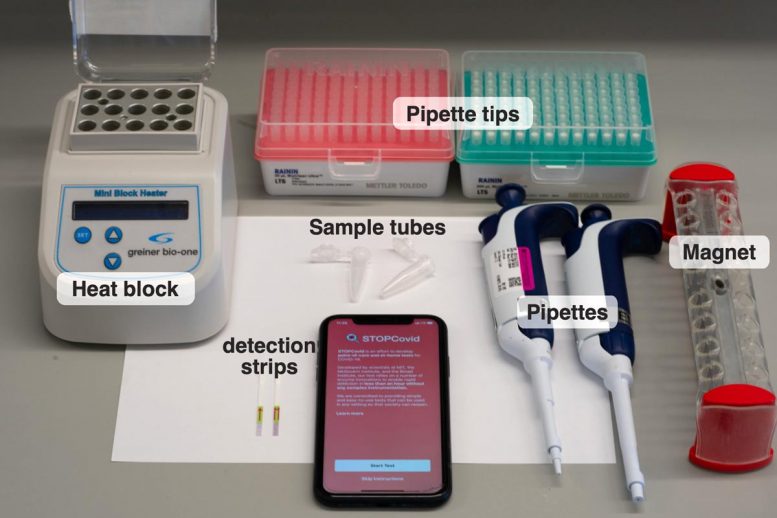
This picture reveals the devices and consumables required for running STOPCovid. Credit: Courtesy of the scientists
In the brand-new variation of STOPCovid reported today, the scientists integrated a procedure to focus the viral hereditary product in a client sample by including magnetic beads that bring in RNA, removing the requirement for pricey filtration packages that are time-intensive and can be in brief supply due to high need. This concentration action improved the test’s level of sensitivity so that it now approaches that of PCR.
“Once we got the viral genomes onto the beads, we found that that could get us to very high levels of sensitivity,” Gootenberg states.
Working with partners Keith Jerome at Fred Hutchinson Cancer Research Center and Alex Greninger at the University of Washington, the scientists checked STOPCovid on 402 client samples — 202 favorable and 200 unfavorable — and discovered that the brand-new test discovered 93 percent of the favorable cases as identified by the basic CDC PCR test.
“Seeing STOPCovid working on actual patient samples was really gratifying,” Ladha states.
They likewise revealed, dealing with Ann Woolley and Deb Hung at Brigham and Women’s Hospital, that the STOPCovid test deals with samples taken utilizing the less intrusive anterior nares swab. They are now evaluating it with saliva samples, which might make at-home tests even simpler to carry out. The scientists are continuing to establish the test with the hope of providing it to end users to assist battle the COVID-19 pandemic.
“The goal is to make this test easy to use and sensitive, so that we can tell whether or not someone is carrying the virus as early as possible,” Zhang states.
Reference: “Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing” by Julia Joung, B.S.; Alim Ladha, B.S.; Makoto Saito, Ph.D.; Nam-Gyun Kim, Ph.D.; Ann E. Woolley, M.D., M.P.H.; Michael Segel, Ph.D.; Robert P.J. Barretto, Ph.D.; Amardeep Ranu, B.S.; Rhiannon K. Macrae, Ph.D.; Guilhem Faure, Ph.D.; Eleonora I. Ioannidi, B.S.; Rohan N. Krajeski, B.S.; Robert Bruneau, B.S.; Meei-Li W. Huang, Ph.D.; Xu G. Yu, M.D.; Jonathan Z. Li, M.D.; Bruce D. Walker, M.D.; Deborah T. Hung, M.D., Ph.D.; Alexander L. Greninger, M.D., Ph.D.; Keith R. Jerome, M.D., Ph.D.; Jonathan S. Gootenberg, Ph.D.; Omar O. Abudayyeh, Ph.D. and Feng Zhang, Ph.D., 16 September 2020, New England Journal of Medicine.
DOI: 10.1056/NEJMc2026172
The research study was moneyed by the National Institutes of Health, the Swiss National Science Foundation, the Patrick J. McGovern Foundation, the McGovern Institute for Brain Research, the Massachusetts Consortium on Pathogen Readiness Evergrande Covid-19 Response Fund, the Mathers Foundation, the Howard Hughes Medical Institute, the Open Philanthropy Project, J. and P. Poitras, and R. Metcalfe.