Computational molecular physics modeling of COVID-19 contaminating the human cell. Credit: Lucy Fallon, Laufer Center, Stony Brook University
New computational tools established at Stony Brook University assistance identify protein structures and determine brand-new treatments for COVID-19.
In the last 5 years, we’ve discovered a lot about the secret lives of proteins — how they work, what they engage with, the equipment that makes them function — and the speed of discovery is speeding up.
The very first three-dimensional protein structure started emerging in the 1970s. Today, the Protein Data Bank, an around the world repository of info about the 3D structures of big biological particles, knows about numerous countless proteins. Just today, the business DeepMind surprised the protein structure world with its precise, AI-driven forecasts.
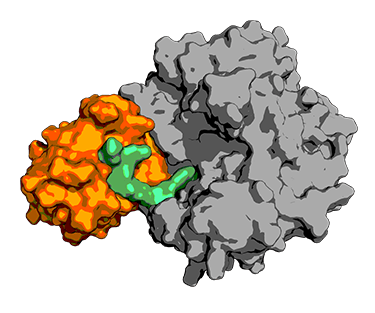
PROTAC particles (green) are artificial particles that string together a protein from the ubiquitin ligase household (gray) with a target protein (orange). Ubiquitin ligases task is to identify other proteins (generally old broken proteins) for damage by the cell equipment. PROTAC particles permit scientists to exploit this equipment to target cancer or infection-related proteins, opening in concept brand-new healing technique to lots of illness. Credit: Emiliano Brini, Laufer Center
But the 3D structure is typically insufficient to really comprehend what a protein depends on, discusses Ken Dill, director of the Laufer Center for Physical and Quantitative Biology at Stony Brook University and a member of the National Academy of Sciences. “It’s like somebody asking how an automobile works, and a mechanic opening the hood of a car and saying, ‘see, there’s the engine, that’s how it works.’”
In the stepping in years, computer system simulations have actually built on and contributed to the understanding of protein habits by setting these 3D molecular devices in movement. Analyzing their energy landscapes, interactions, and characteristics has actually taught us a lot more about these prime movers of life.
“We’re really trying to ask the question: how does it work? Not just, how does it look?” Dill stated. “That’s the essence of why you want to know protein structures in the first place, and one of the biggest applications of this is for drug discovery.”
Writing in Science publication in November 2020, Dill and his Stony Brook coworkers Carlos Simmerling and Emiliano Brini shared their viewpoints on the development of the field.
“Computational Molecular Physics is an increasingly powerful tool for telling the stories of protein molecule actions,” they composed. “Systematic enhancements in forcefields, boosted tasting techniques, and accelerators have actually made it possible for [computational molecular physics] to reach timescales of essential biological actions…. At this rate, in the next quarter century, we’ll be informing stories of protein particles over the entire life expectancy, 10s of minutes, of a bacterial cell.”
Speeding Simulations
Decades after the very first vibrant designs of proteins, nevertheless, computational biophysicists still deal with significant difficulties. To work, simulations require to be precise; and to be precise, simulation requires to advance atom by atom and femtosecond (10^-12 seconds) by femtosecond. To match the timescales that matter, simulations should cross split seconds or milliseconds — that is, countless time-steps.
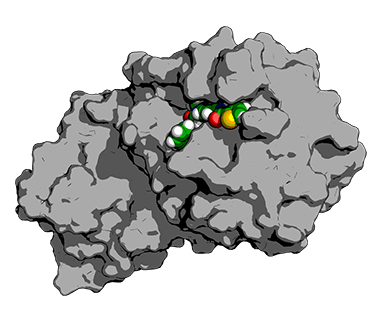
The forecast by the Laufer Center group of the binding present of a little drug like particle (green) to its target protein (gray). The forecasted structure is significanlty comparable to the experimentally observed one. Credit: Emiliano Brini, Laufer Center
“Computational molecular physics has developed at a fast clip relatively speaking, but not enough to get us into the time and size and motion range we need to see,” he stated.
One of the primary techniques scientists utilize to comprehend proteins in this method is called molecular characteristics. Since 2015, with assistance from the National Institutes of Health and the National Science Foundation, Dill and his group have actually been working to accelerate molecular characteristics simulations. Their approach, called MELD, speeds up the procedure by supplying unclear however essential info about the system being studied.
Dill compares the approach to a witch hunt. Instead of asking somebody to discover a treasure that might be anywhere, they supply a map with hints, stating: ‘it’s either near Chicago or Idaho.’ In the case of real proteins, that may imply informing the simulation that a person part of a chain of amino acids is near another part of the chain. This constricting of the search field can accelerate simulations considerably — often more than 1000-times quicker — making it possible for unique research studies and supplying brand-new insights.
Protein Structure Predictions for COVID-19
One of the most essential usages of biophysical modeling in our lives is drug discovery and advancement. 3D designs of infections or germs assist determine vulnerable points in their defenses, and molecular characteristics simulations identify what little particles might bind to those opponents and gum up their works without needing to check every possibility in the laboratory.
Dill’s Laufer Center group is associated with a variety of efforts to discover drugs and treatments for COVID-19, with assistance from the White House-arranged COVID-19 HPC Consortium, an effort amongst Federal federal government, market, and scholastic leaders to supply access to the world’s most effective high-performance computing resources in assistance of COVID-19 research study.
“Everyone dropped other things to work on COVID-19,” Dill remembered.
The primary step the group took was to utilize MELD to identify the 3D shape of the coronavirus’ unidentified proteins. Only 3 of the 29 of the infection’ proteins have actually been definitively fixed up until now. “Most structures are not known, which is not a good beginning for drug discovery,” he stated. “Can we predict structures that are not known? That’s the primary thing that we used Frontera for.”
The Frontera supercomputer at the Texas Advanced Computing Center (TACC) — the fastest at any university worldwide — permitted Dill and his group to make structure forecasts for 19 extra proteins. Each of these might work as an opportunity for brand-new drug advancements. They have actually made their structure forecasts openly offered and are dealing with groups to experimentally check their precision.
While it appears like the vaccine race is currently near stating a winner, the preliminary of vaccines, drugs, and treatments are just the beginning point for a healing. As with HIV, it is most likely that the very first drugs established will not deal with all individuals, or will be exceeded by more reliable ones with less side-effects in the future.
Dill and his Laufer Center group are playing the long video game, wanting to discover targets and systems that are more appealing than those currently being established.
Repurposing Drugs and Exploring New Approaches
A 2nd job by the Laufer Center group utilizes Frontera to scan countless commercially offered little particles for effectiveness versus COVID-19, in partnership with Dima Kozakov’s group at Stony Brook University.
“By focusing on the repurposing of commercially available molecules it’s possible, in principle, to shorten the time it takes to find a new drug,” he stated. “Kozakov’s group has the ability to quickly screen thousands of molecules to identify the best hundred ones. We use our physics modeling to filter this pool of candidates even further, narrowing the options experimentalists need to test.”
A 3rd job is studying an intriguing cellular protein referred to as PROTAC that directs the “trash collector proteins” of human cells to get particular target proteins that they would not generally get rid of.
“Our cell has smart ways to identify proteins that needs to be destroyed. It gets next to it, puts a sticker on it, and the proteins who collect trash take it away,” he described. “Initially PROTAC particles have actually been utilized to target cancer associated proteins. Now there is a push to move this idea to target SARS-CoV-2 proteins.”
Collaborating with Stony Brook chemist Peter Tonge, they are working to replicate the interaction of unique PROTACS with the COVID-19 infection. “These are some of our most ambitious simulations, both in term of the size of the systems we are tackling and in terms of the chemical complexity,” he stated. “Frontera is a crucial resource to give us sufficient turnaround times. For one simulation we need 30 GPUs and four to five days of continuous calculations.”
The group is establishing and checking their procedures on a non-COVID test system to standard their forecasts. Once they choose a procedure, they will use this style treatment to COVID systems.
Every protein has a story to inform and Dill, Brini and their partners are developing and using the tools that assist illuminate these stories. “There are some problems in protein science where we believe the real challenge is getting the physics and math right,” Dill concluded. “We’re testing that hypothesis on COVID-19.”
Reference: “Protein storytelling through physics” by Emiliano Brini, Carlos Simmerling and Ken Dill, 27 November 2020, Science.
DOI: 10.1126/science.aaz3041





