The advancement of mRNA knock-down innovations for usage in vertebrate organisms such as zebrafish has actually been restricted. CRISPR-RfxCas13d has actually been determined as a reliable technique for the organized research study of gene function in vivo throughout embryogenesis throughout a variety of animal types. Credit: Bazzini Lab, Stowers Institute
CRISPR is a gene-editing tool that has actually allowed scientists to study the function of various genes in design organisms. Scientists at the Stowers Institute for Medical Research in Kansas City, Missouri, and the Andalusian Center of Developmental Biology at Pablo de Olavide University in Seville, Spain, have actually now utilized this effective innovation to target gene messages in animal design embryos to acquire a much better understanding of the hereditary programs that direct early phases of vertebrate advancement.
In a report released online today (August 7, 2020) in Developmental Cell, the Stowers group, led by Assistant Investigator Ariel Bazzini, PhD, and partners reveal that the innovation, called CRISPR-Cas13, has the ability to target RNA — DNA’s chemical cousin that brings messages required to construct proteins — in embryonic animal designs in a particular and organized way, permitting scientists to study the function of RNA in the earliest hours of advancement. The scientists reveal that the CRISPR-Cas13 technique works in zebrafish, killifish, medaka, and mouse embryos, and therefore might be utilized to check out early developmental hereditary programs in a broad variety of animal types.
Since its discovery, researchers have actually been utilizing the much-lauded gene modifying tool CRISPR to change the DNA of design organisms and discover the functions of countless genes. Now, scientists at the Stowers Institute for Medical Research in Kansas City, Missouri, and the Andalusian Center of Developmental Biology at Pablo de Olavide University in Seville, Spain, have actually utilized the innovation to target gene messages (messenger RNA) associated with early vertebrate advancement.
By interfering with gene messages (RNA) rather of the hidden genes (DNA), scientists can study genes that may formerly have actually been challenging or difficult to control due to the fact that they were necessary to life or associated with an important phase of biological advancement. This method likewise enables targeting of maternally-contributed gene RNAs, which are transferred in the egg to begin the earliest hereditary programs.
The research study, which appears online August 7, 2020, in the journal Developmental Cell, develops using CRISPR-Cas innovation to target RNA in embryonic animal designs in a particular and organized way. The findings show the strategy can be used to a broad variety of water and terrestrial designs consisting of zebrafish, medaka, killifish, and mice.
“The exciting thing about this study is not just what we found, but what we can do,” states Ariel Bazzini, PhD, an assistant detective at the Stowers Institute and co-leader of the research study. “We still don’t understand how genes jumpstart the earliest stages of development. Now we can find out by targeting their RNA messages, one by one.”
“We are also very excited about the low cost of the technique,” Bazzini states. “Any lab working with zebrafish or other animal embryos could use this method. Indeed, we have already distributed the reagents and protocol to several labs around the world.”
Before advancement even starts, egg should initially satisfy sperm. The resulting embryo brings half the genes from the mom and half from the dad. In addition to its genome, the embryo has parts such as RNA and proteins offered by the mom.
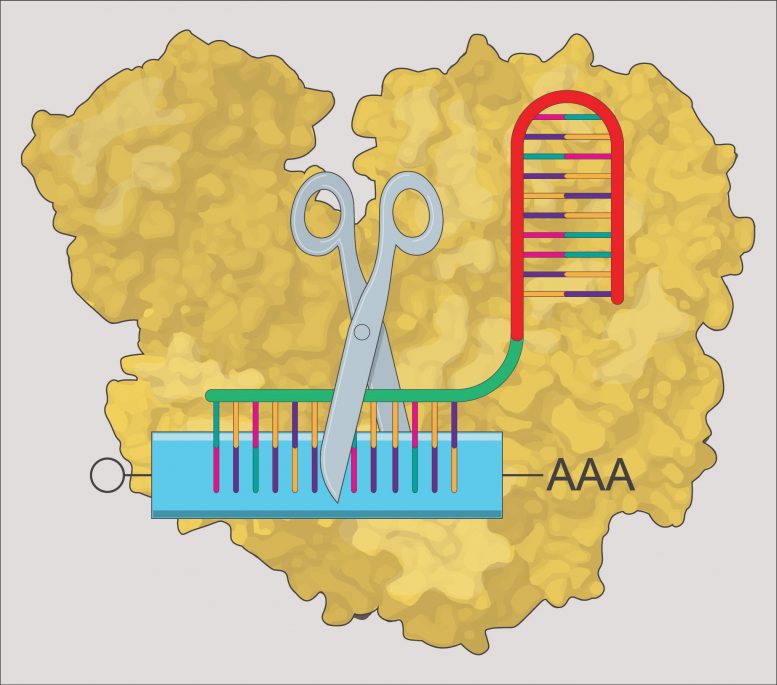
The advancement of mRNA knock-down innovations for usage in vertebrate organisms such as zebrafish has actually been restricted. CRISPR-RfxCas13d has actually been determined as a reliable technique for the organized research study of gene function in vivo throughout embryogenesis throughout a variety of animal types. Credit: Bazzini Lab, Stowers Institute
“That maternal contribution is a mystery that many of us want to solve,” states Bazzini. However, tries to methodically target RNA in zebrafish, the design organism of option for numerous developmental biologists, have actually been not successful. The aptly-named RNA disturbance technique, which has actually been a pillar in research studies of gene function, does not operate in zebrafish, or other fish or frogs. Other techniques utilizing synthetics strips of hereditary code referred to as morpholinos or antisense oligonucleotides have actually in some cases been related to toxicity and off-target impacts.
So when Bazzini and his partner and pal Miguel A. Moreno-Mateos, PhD, a teacher at Pablo de Olavide University, saw reports that CRISPR innovation had actually been utilized to deteriorate RNA in yeast, plants, and mammalian cells, they aspired to provide it a shot. Moreno-Mateos was a postdoc in Antonio Giraldez’s laboratory at Yale University at the exact same time as Bazzini, and is thought about a professional on the optimization of CRISPR-Cas innovation in vivo.
The CRISPR-Cas13 system depends upon 2 components — a brief RNA series referred to as a “guide” RNA, and an enzyme called Cas13 (part of the Cas, or CRISPR-associated, household of proteins) that cuts any RNA messages in the cell that might line up and bind to that guide series. The scientists evaluated 4 various Cas13 proteins that had actually been effectively utilized in previous research studies. They discovered that the Cas13 proteins were either ineffective or hazardous to the establishing zebrafish, other than for one protein, called RfxCas13d.
They then took a look at whether targeting RNA with CRISPR-RfxCas13d in zebrafish embryos might recreate the exact same flaws as modifying the organism’s underlying DNA. For example, when they targeted the RNA of the tbxta gene, which is required for growing a tail, the zebrafish embryos were tailless.
The scientists went on to reveal that the CRISPR system might effectively target a range of RNAs, both those offered by the mom in addition to those produced by the embryo, reducing RNA levels by approximately 76%. Collaborators within and beyond Stowers assisted obtain that fact, and revealed that the strategy likewise operates in killifish, medaka, and mouse embryos.
“The CRISPR-RfxCas13d system is an efficient, specific and inexpensive method that can be used in animal embryos in a comprehensive manner,” states Moreno-Mateos, who is likewise co-leader of the research study. “With this tool we will help to understand fundamental questions in biology and biomedicine.”
One of the basic concerns the scientists wish to pursue is the function that RNA plays in the earliest hours of advancement. The RNAs left by the mom need to be gotten rid of at specifically the exact same time that the genome of the embryo comes online; otherwise, the embryo never ever establishes.
“We think this tool could have a profound effect on our understanding of infertility and developmental problems in general,” states Bazzini.
“The Stowers facilities and collaborative environment have allowed us to test CRISPR technology in other animal model systems,” Bazzini states. “When I joined Stowers about four years ago, I would have never predicted that my lab would be doing experiments in mouse or killifish models. It’s been a fun adventure!”
Reference: 7 August 2020, Developmental Cell.
Other coauthors from the Stowers Institute consist of Gopal Kushawah, PhD, Michelle DeVore, Huzaifa Hassan, Wei Wang, PhD, Timothy J. Corbin, Andrea M. Moran, and Alejandro Sánchez Alvarado, PhD.
This research study was moneyed by the Stowers Institute for Medical Research, Pablo de Olavide University, Consejo Superior de Investigaciones Cientificas, and Junta de Andalucia. Additional assistance consisted of the Ramon y Cajal program (RyC-2017-23041) and grants (BFU2017-86339-P, PGC2018-097260-B-I00, and MDM-2016-0687) from the Spanish Ministerio de Ciencia, Innovación y Universidades; the Springboard program from Centro Andaluz de Biología del Desarrollo; Genome Engineer Innovation 2019 Grant from Synthego; the Pew Innovation Fund; Innovate Peru (grant 168-PNICP-PIAP-2015); and FONDECYT (take a trip grant 043-2019).





