The Gram-unfavorable cell envelope and paths of drug fluxes throughout it. Credit: Zhao, S., Adamiak, J.W., Bonifay, V. et al. Defining brand-new chemical area for drug penetration into Gram-unfavorable germs. Nat Chem Biol 16, 1293-1302 (2020). DOI 10.1038/s41589-020-00674-6
University of Oklahoma scientists released a viewpoint post in the journal Nature Chemical Biology that resolves the space in the discovery of brand-new prescription antibiotics.
“The continuing emergence of antibiotic-resistant bacteria, and our inability to develop new antibiotics to combat them, represent two of the most wicked healthcare problems we face,” stated OU vice president for research study and collaborations Tomás Díaz de la Rubia. “OU’s team is working hard to develop solutions to these major challenges, and their opinion article helps bring visibility and attention to these issues at a time when it’s needed most.”
The research study group is made up of OU scientists Helen Zgurskaya, Valentin Rybenkov and Adam Duerfeldt, all in the Department of Chemistry and Biochemistry, College of Arts and Sciences, with scientists from the Memorial Sloan Kettering Cancer Center, led by Derek Tan, and business scientists from Merck & Co.
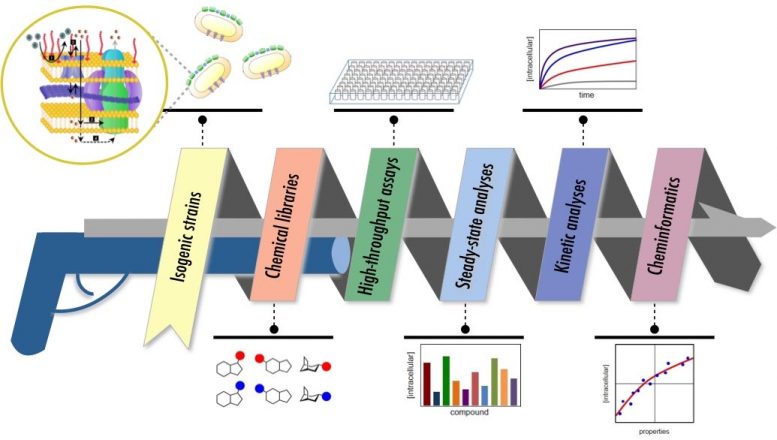
Comprehensive technique to establishing cheminformatic tools to forecast Gram-unfavorable bacterial substance build-up. Credit: Justyna Adamiak
“The rapid spread of antibiotic-resistant bacteria in clinics challenges our modern medicine and the traditional approaches to antibiotic discovery fail to generate new drugs needed for treatment of antibiotic resistant infections,” Zgurskaya stated. “The existing COVID-19 pandemic even more amplifies this issue since clients in extensive care systems are especially susceptible to such infections … (our) group is dealing with establishing brand-new tools to assist the discovery and optimization of brand-new anti-bacterial representatives.”
Zgurskaya includes that the increasing frequency of antibiotic resistance has actually produced a substantial healthcare difficulty and will gradually get worse without ingenious options.
“In particular, gram-negative pathogens present both biological and chemical challenges that hinder the discovery of new antibacterial drugs,” Zgurskaya stated. “As a result of these challenges, intensive screening campaigns have led to few successes, highlighting the need for new approaches to identify regions of chemical space that are specifically relevant to antibacterial drug discovery.”
In the post, the research study group offers a summary of emerging insights into this issue and detail a basic technique for scientists and researchers to resolve it.
“The overall goal is to develop robust cheminformatic tools to predict gram-negative permeation and efflux, which can then be used to guide medicinal chemistry campaigns and the design of antibacterial discovery libraries,” Zgurskaya stated.
Reference: “Defining new chemical space for drug penetration into Gram-negative bacteria” by Shibin Zhao, Justyna W. Adamiak, Vincent Bonifay, Jitender Mehla, Helen I. Zgurskaya and Derek S. Tan, 16 November 2020, Nature Chemical Biology.
DOI: 10.1038/s41589-020-00674-6
The research study was supported with financing from the U.S. Department of Health and Human Services and the National Institutes of Health. The post, Defining brand-new chemical area for drug penetration into Gram-unfavorable germs, is readily available in the November 2020 problem of the scholastic journal, Nature Chemical Biology.





