An global group has actually established the most thorough bird ancestral tree, covering 93 million years and 92% of bird households, utilizing innovative computational techniques and supercomputing resources. Credit: SciTechDaily.com
An international group of scientists has actually built the most thorough and substantial bird ancestral tree to date, detailing the evolutionary connections amongst 363 bird < period class ="glossaryLink" aria-describedby ="tt" data-cmtooltip ="<div class=glossaryItemTitle>species</div><div class=glossaryItemBody>A species is a group of living organisms that share a set of common characteristics and are able to breed and produce fertile offspring. The concept of a species is important in biology as it is used to classify and organize the diversity of life. There are different ways to define a species, but the most widely accepted one is the biological species concept, which defines a species as a group of organisms that can interbreed and produce viable offspring in nature. This definition is widely used in evolutionary biology and ecology to identify and classify living organisms.</div>" data-gt-translate-attributes="[{"attribute":"data-cmtooltip", "format":"html"}]" tabindex ="0" function ="link" > types over93 million years.This chart represents 92 % of all bird households.
The advance was enabled in big part thanks to advanced computational techniques established by engineers at the(****************************************************************************************************************************************************** )ofCaliforniaSanDiego, integrated with the university’s cutting edge supercomputing resources at theSanDiegoSupercomputerCenterThese innovations have actually allowed scientists to evaluate huge quantities of genomic information with high< period class ="glossaryLink" aria-describedby ="tt" data-cmtooltip ="<div class=glossaryItemTitle>accuracy</div><div class=glossaryItemBody>How close the measured value conforms to the correct value.</div>" data-gt-translate-attributes =" (** )" tabindex ="0" function ="link" > precision and speed, preparing for the building and construction of the most thorough bird ancestral tree ever put together.
The advance is detailed in 2 complementary documents released onApril 1 inNature and theProceedings of theNationalAcademy ofSciences( PNAS)The upgraded ancestral tree, reported inNature, exposed patterns in the evolutionary history of birds following the catastrophic mass termination occasion that eliminated the dinosaurs66 million years back.
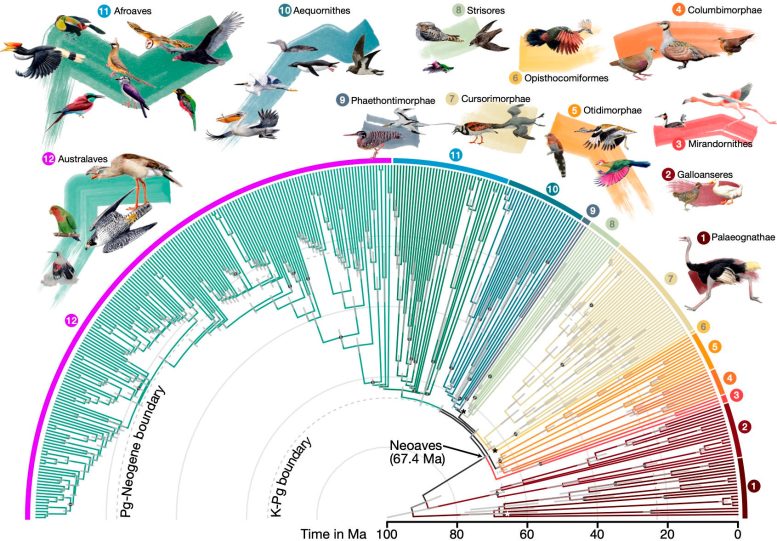
The upgraded bird ancestral tree, released inNature, marking93 million years of evolutionary relationships in between363 bird types.Credit:JonFjelds å( illustrations) andJosefinStiller
(****************************************************************************************************************************************************************************************** )observed sharp boosts in efficient population size, alternative rates, and relative brain size in early risers, shedding brand-new light on the adaptive systems that drove bird diversity in the consequences of this critical occasion. In the buddy paper released in PNAS, scientists carefully analyzed among the branches of the brand-new ancestral tree and discovered that flamingos and doves are more distantly associated than previous genome-wide analyses had actually revealed.
The work belongs to the Bird 10,000 Genomes (B10 K) Project, a multi-institutional effort led by University of Copenhagen, Zhejiang University, and UC San Diego that intends to create draft genome series for about 10,500 extant bird types.
“Our goal is to reconstruct the entire evolutionary history of all birds,” stated Siavash Mirarab, teacher of electrical and computer system engineering at the UC San Diego Jacobs School of Engineering, who is a co-senior author on the Nature paper, along with very first and co-corresponding author on the PNAS paper.
Piecing together the past
At the heart of these research studies lies a suite of algorithms called ASTRAL, which Mirarab’s laboratory established to presume evolutionary relationships with unmatched scalability, precision, and speed. By utilizing the power of these algorithms, the group incorporated genomic information from over 60,000 genomic areas, offering a robust analytical structure for their analyses.
The scientists then analyzed the evolutionary history of specific sections throughout the genome. From there, they pieced together a mosaic of gene trees, which were then assembled into a thorough types tree. This careful method allowed the scientists to build a brand-new and enhanced bird ancestral tree that marks intricate branching occasions with impressive accuracy and information, even in cases of historic unpredictability.
“We found that our method of adding tens of thousands of genes to our analysis was actually necessary to resolve evolutionary relationships between bird species,” statedMirarab “You really need all that genomic data to recover what happened in this certain period of time 65-67 million years ago with high confidence.”

In the research study released in PNAS, scientists carefully analyzed among the branches of the upgraded bird ancestral tree and discovered that groups consisting of flamingos and doves are more distantly associated than previous genome-wide analyses had actually revealed and associated the outcome to an uncommon area of chromosome 4. Credit: Ed Braun (illustrations), Daniel J. Field (bird images) and Siavash Miarab
The group’s capability to carry out these analyses on enormous datasets was enabled since Mirarab’s laboratory developed their computational techniques to work on effective GPU devices. They ran their computations on the Expanse supercomputer at the San Diego Supercomputer at UC San Diego.
“We were fortunate to have access to such a high-end supercomputer,” statedMirarab “Without Expanse, we would not have actually had the ability to run and rerun our analyses on such big datasets in a sensible quantity of time.”
The scientists likewise took a look at the impacts of various genome tasting techniques on the precision of the tree. They revealed that 2 methods– sequencing numerous genes from each types, along with sequencing numerous types– integrated together are very important for rebuilding this evolutionary history.
“Because we used a mixture of both strategies, we could test which approach has stronger impacts on phylogenetic reconstruction,” stated Josefin Stiller, teacher of biology at the University of Copenhagen and lead author of the Nature paper. “We found that it was more important to sample many genetic sequences from each organism than it was to sample from a broader range of species, although the latter method helped us to date when different groups evolved.”
Correcting the past
With the help of their innovative computational techniques, the scientists were likewise able to clarify something uncommon that they had actually found in among their previous research studies: a specific area of one chromosome in the bird genome had actually stayed the same for countless years, devoid of the anticipated patterns of hereditary recombination.
This anomaly at first led the scientists to improperly group flamingos and doves together as evolutionary cousins, for they appeared carefully associated based upon this the same area of < period class ="glossaryLink" aria-describedby ="tt" data-cmtooltip ="<div class=glossaryItemTitle>DNA</div><div class=glossaryItemBody>DNA, or deoxyribonucleic acid, is a molecule composed of two long strands of nucleotides that coil around each other to form a double helix. It is the hereditary material in humans and almost all other organisms that carries genetic instructions for development, functioning, growth, and reproduction. Nearly every cell in a person’s body has the same DNA. Most DNA is located in the cell nucleus (where it is called nuclear DNA), but a small amount of DNA can also be found in the mitochondria (where it is called mitochondrial DNA or mtDNA).</div>" data-gt-translate-attributes="[{"attribute":"data-cmtooltip", "format":"html"}]" tabindex ="0" function ="link" > DNAThat’s since their previous analysis was based upon the genomes of(************************************************************************************************************************** )bird types. But by duplicating their analysis utilizing the genomes of363 types, a more precise ancestral tree emerged that moved doves even more from flamingos.Moreover, utilizing 6 premium genomes offered by theVertebrateGenomeProject( VGP)– led by co-authorErich(********************************************************************************************************************************************************************************************************************************************* )a teacher of neurobiology atRockefellerUniversity–Mirarab and coworkers had the ability to find and putatively discuss this unexpected pattern.
“What’s surprising is that this period of suppressed recombination could mislead the analysis,” statedEdwardBraun, teacher of biology at the < period class ="glossaryLink" aria-describedby ="tt" data-cmtooltip ="<div class=glossaryItemTitle>University of Florida</div><div class=glossaryItemBody>Established in 1853, the University of Florida (Florida or UF) is a public land-grant, sea-grant, and space-grant research university in Gainesville, Florida. It is home to 16 academic colleges and more than 150 research centers and institutes. University of Florida offers multiple graduate professional programs, including business administration, engineering, law, dentistry, medicine, pharmacy, and veterinary medicine, and administers 123 master's degree programs and 76 doctoral degree programs in eighty-seven schools and departments.</div>" data-gt-translate-attributes="[{"attribute":"data-cmtooltip", "format":"html"}]" tabindex ="0" function ="link" >(****************************************************************************************************************************************************** )ofFlorida and co-corresponding author of the PNAS paper. “And because it could mislead the analysis, it was actually detectable more than 60 million years in the future. That’s the cool part.”
Next actions
The effect of this work extends far beyond studying the evolutionary history of birds. The computational techniques originated by Mirarab’s laboratory have actually turned into one of the requirement tools for rebuilding evolutionary trees for a range of other animals.
Moving forward, the group is continuing their efforts to build a total photo of bird development. Biologists are dealing with sequencing the genomes of extra bird types in the hopes of broadening the ancestral tree to consist of countless bird genera. Meanwhile, computational researchers led by Mirarab are improving their algorithms to accommodate even bigger datasets to make sure that analyses in future research studies are carried out with high speed and precision.
Reference: “Complexity of avian evolution revealed by family-level genomes” by Josefin Stiller, Shaohong Feng, Al-Aabid Chowdhury, Iker Rivas-Gonz ález, David A. Duch êne, Qi Fang, Yuan Deng, Alexey Kozlov, Alexandros Stamatakis, Santiago Claramunt, Jacqueline M. T. Nguyen, Simon Y. W. Ho, Brant C. Faircloth, Julia Haag, Peter Houde, Joel Cracraft, Metin Balaban, Uyen Mai, Guangji Chen, Rongsheng Gao, Chengran Zhou, Yulong Xie, Zijian Huang, Zhen Cao, Zhi Yan, Huw A. Ogilvie, Luay Nakhleh, Bent Lindow, Benoit Morel, Jon Fjelds å, Peter A. Hosner, Rute R. da Fonseca, Bent Petersen, Joseph A. Tobias, Tam ás Sz ékely, Jonathan David Kennedy, Andrew Hart Reeve, Andras Liker, Martin Stervander, Agostinho Antunes, Dieter Thomas Tietze, Mads Bertelsen, Fumin Lei, Carsten Rahbek, Gary R. Graves, Mikkel H. Schierup, Tandy Warnow, Edward L. Braun, M. Thomas P. Gilbert, Erich D. Jarvis, Siavash Mirarab and Guojie Zhang, 1 April 2024, Nature
DOI: 10.1038/ s41586-024-07323 -1
“A region of suppressed recombination misleads neoavian phylogenomics” by Siavash Mirarab, Iker Rivas-Gonz ález, Shaohong Feng, Josefin Stiller, Qi Fang, Uyen Mai, Glenn Hickey, Guangji Chen, Nadolina Brajuka, Olivier Fedrigo, Giulio Formenti, Jochen B. W. Wolf, Kerstin Howe, Agostinho Antunes, Mikkel H. Schierup, Benedict Paten, Erich D. Jarvis, Guojie Zhang and Edward L. Braun, 1 April 2024, Proceedings of the National Academy of Sciences
DOI: 10.1073/ pnas.2319506121





