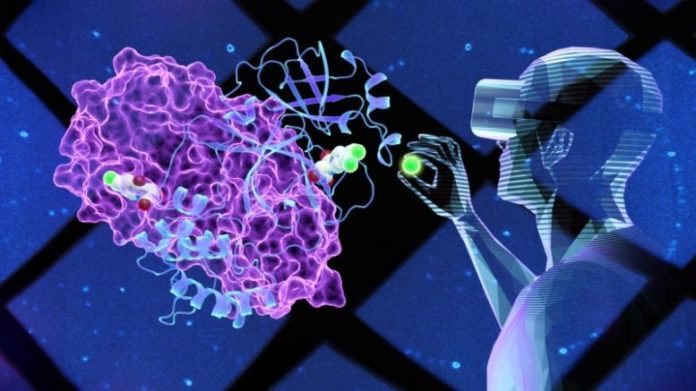
Virtual truth innovation allowed researchers to look “inside” the Covid-19 infection and establish an unique particle that can prevent its primary protease enzyme. Credit: Jill Hemman/ ORNL
Virtual truth (VR) innovation makes it possible for researchers to produce 3-D designs of an item and after that essentially go “inside” to take a look around to much better comprehend its structure and function.
This is what scientists at the Department of Energy’s (DOE’s) Oak Ridge National Laboratory (ORNL) did to study the SARS-CoV-2 infection that triggered the COVID-19 pandemic. The group utilized neutrons and x-rays to map part of the internal structure of the coronavirus to produce a precise 3-D design. Specifically, the researchers mapped the primary protease (Mpro), an enzyme associated with the infection duplication, to which they had actually included an initial little particle found utilizing high-speed computer system screening.
Using VR to take a look at the enzyme design, the researchers essentially built various little particles by customizing their structures to see if any freshly developed substances might fit, or bind, to an essential website on the Mpro enzyme surface area. A strong adequate binding might prevent, or block, the enzyme from working, which is crucial to stopping the infection from increasing in clients with COVID-19
To identify the results of particular chemical adjustments on how firmly the 19 inhibitor prospects bind to the Mpro enzyme, the group manufactured each inhibitor particle and determined their binding strengths. The more powerful the binding, the better the inhibitor would obstruct the enzyme from working and the infection from duplicating.
One of the test inhibitors, identified HL-3-68, showed a remarkable capability to bind to and prevent the function of Mpro compared to others that were checked. Details of the research study, entitled “Structural, electronic and electrostatic determinants for inhibitor binding to subsites S1 and S2 in SARS-CoV-2 main protease,” are released in the Journal of Medicinal Chemistry
“Our study was designed to better understand how molecules bind at the active site of the Mpro enzyme, which plays a key role in SARS-CoV-2 replication,” stated lead author DanielKneller “In the procedure of checking the particles we developed, we found one consisting of a single additional chlorine atom that revealed a higher capability to preventMpro This unique chemical structure is various than what has actually been formerly studied by the international neighborhood and might open brand-new opportunities of research study with amazing possibilities for combating SARS-CoV-2.”
The active website on the Mpro enzyme prevails to other kinds of coronaviruses and does not appear to quickly alter– providing a chance to potentially create an antiviral treatment that works versus several SARS-CoV-2 versions and other coronaviruses.
Equally crucial is that the active website is various from those understood in human enzymes, which would lessen the capacity for unexpected binding that might result in negative effects in clients.
The x-ray measurements and production of the Mpro enzyme samples were carried out by the Center for Structural and Molecular Biology utilizing centers at ORNL’s Spallation Neutron Source (SNS) and resources at the High Flux Isotope Reactor (HFIR). The inhibitor prospects were manufactured by co-authors Hui Li and Peter Bonnesen of the Macromolecular Nanomaterials group at the Center for Nanophase Materials Sciences (CNMS).
“This study combined a plethora of biophysical, biochemical and molecular biology methods, and included virtual reality-assisted structural analysis and small molecule building, bringing together scientists from across ORNL, Argonne National Laboratory, the National Institutes of Health, and the University of Tennessee-Knoxville. The collaborative nature of the study allowed us to uncover the rules small molecule inhibitors must obey when binding to the enzyme in order to be useful for further steps in the long process of drug design and development,” stated matching author Andrey Kovalevsky.
Added co-corresponding author Peter Bonnesen, “this was a new and exciting project for the CNMS to work on, and it drew on our expertise in synthesizing custom organic molecules for our users. For this project, we provided our SNS colleagues with a couple of candidate molecules at a time. As results came back regarding the molecules’ effectiveness as inhibitors, the team would discuss what adjustments to make to the molecular structure. Then Hui and I would go back to the lab to make these new candidate inhibitors.”
The research study likewise clarified the Mpro enzyme’s capability to alter its shape and alter its electrical charge from favorable to unfavorable, or from unfavorable to favorable, according to the size and structure of the inhibitor particle it binds to. These functions are essential to comprehend when establishing a reliable inhibitor particle.
For the neutron spreading research study, the researchers utilized the macromolecular neutron diffractometer (MaNDi) at the SNS for its capability to gather information from the relatively little samples the group needed to deal with.
“Due to the pressing nature of research related to the SARS-CoV-2 virus, we were only able to grow relatively small samples of the Mpro enzyme,” stated co-author LeightonCoates “As smaller samples scatter neutrons weakly and this results in “noisy” neutron information, information analysis can be tough. The time structure of the neutron beam at the MaNDi instrument enabled us to eliminate the majority of the sound, consequently increasing the signal-to-noise ratio, providing us a lot more helpful information to deal with.”
Next actions for the ORNL scientists consist of screening chemical adjustments of the HL-3-68 inhibitor to identify if any freshly developed substances can bind even much better than HL-3-68 to better prevent the Mpro enzyme and eventually avoid the coronavirus from duplicating.
Meanwhile, the scientists made their information openly readily available by means of the Protein Data Bank to speed up notifying and helping the world’s clinical and medical neighborhoods. Of course, more research study and tests are required to verify the efficiency and security of any inhibitor as a COVID-19 treatment. However, this research study might use a chance for other researchers to perform extra research study that would benefit billions of individuals worldwide.
Reference: “Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease” by Daniel W. Kneller, Hui Li, Stephanie Galanie, Gwyndalyn Phillips, Audrey Labb é, Kevin L. Weiss, Qiu Zhang, Mark A. Arnould, Austin Clyde, Heng Ma, Arvind Ramanathan, Colleen B. Jonsson, Martha S. Head, Leighton Coates, John M. Louis, Peter V. Bonnesen and Andrey Kovalevsky, 27 October 2021, Journal of Medicinal Chemistry
DOI: 10.1021/ acs.jmedchem.1 c01475
The paper’s other co-authors consist of Stephanie Galanie, Gwyndalyn Phillips, Audrey Labb é, Kevin L. Weiss, Qiu Zhang, Mark A. Arnould, Austin Clyde, Heng Ma, Arvind Ramanathan, Colleen B. Jonsson, Martha S. Head, and John M.Louis Hugh O’Neill from ORNL helped throughout sample preparation.
COVID-19 research study at ORNL was supported in part by the Office of Science’s National Virtual Biotechnology Laboratory, a consortium of DOE nationwide labs concentrated on reacting to COVID-19, with financing offered by the Coronavirus CARESAct This work was likewise supported by the National Institutes of Health’s National Institute of Diabetes and Digestive and Kidney Diseases.