By
Mammalian cells that have actually been effectively genetically crafted utilizing the STAMPScreen technique. Credit: Wyss Institute at Harvard University
STAMPScreen Pipeline Helps Streamline Genetic Studies in Mammalian Cells
Today’s hereditary engineers have a huge selection of resources at their disposal: an ever-increasing variety of huge datasets readily available online, extremely accurate gene modifying tools like CRISPR, and inexpensive gene sequencing techniques. But the expansion of brand-new innovations has actually not featured a clear roadmap to assist scientists find out which genes to target, which tools to utilize, and how to translate their outcomes. So, a group of researchers and engineers at Harvard’s Wyss Institute for Biologically Inspired Engineering, Harvard Medical School (HMS), and the MIT Media Lab chose to make one.
The Wyss group has actually produced an integrated pipeline for carrying out hereditary screening research studies, incorporating every action of the procedure from determining target genes of interest to cloning and evaluating them rapidly and effectively. The procedure, called Sequencing- based Target Ascertainment and Modular Perturbation Screening (STAMPScreen), is explained in Cell Reports Methods, and the associated open-source algorithms are readily available on GitHub.
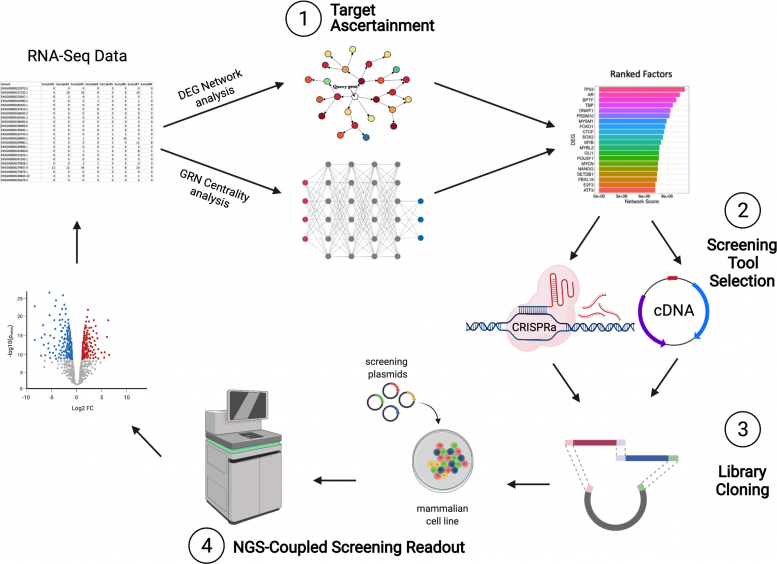
The STAMPScreen workflow is an integrated pipeline that permits scientists to rapidly and quickly evaluate a speculative database for possible genes of interest (1 ), pick which screening tool to utilize (2 ), produce a screening library (3 ), and utilize next-generation sequencing to screen genes in vivo (4 ). The private actions can likewise be utilized in other workflows. Credit: Wyss Institute at Harvard University
“STAMPScreen is a streamlined workflow that makes it easy for researchers to identify genes of interest and perform genetic screens without having to guess which tool to use or what experiments to perform to get the results they want,” stated matching author Pranam Chatterjee,Ph D., a previous college student at the MIT Media Lab who is now the Carlos M. Varsavsky Research Fellow at HMS and the WyssInstitute “It is fully compatible with many existing databases and systems, and we hope that many scientists are able to take advantage of STAMPScreen to save themselves time and improve the quality of their results.”
Frustration is the mom of innovation
Chatterjee and Christian Kramme, a co-first author of the paper, were annoyed. The 2 researchers were attempting to check out the hereditary foundations of various elements of biology– like fertility, aging, and resistance– by integrating the strengths of digital techniques (believe algorithms) and genetic modification (think gene sequencing). But they kept encountering issues with the different tools and procedures they were utilizing, which are prevalent in science laboratories.
The algorithms that supposed to sort through an organism’s genes to determine those with a considerable influence on a provided biological procedure might inform when a gene’s expression pattern altered, however didn’t supply any insight into the reason for that modification. When they wished to check a list of prospect genes in living cells, it wasn’t instantly clear what kind of experiment they ought to run. And a number of the tools readily available to place genes into cells and evaluate them were costly, lengthy, and inflexible.

Co- very first author of the paper, Christian Kramme, at his bench at the WyssInstitute Credit: Wyss Institute at Harvard University
“I was using methods known as Golden Gate and Gateway to clone genes into vectors for screening experiments, and it took me months and thousands of dollars to clone 50 genes. And using Gateway, I couldn’t physically barcode the genes to identify which one got into which vector, which was a crucial requirement for my downstream sequencing-based experimental design. We figured there had to be a better way to do this kind of research, and when we couldn’t find one, we took on the challenge of creating it ourselves,” stated Kramme, who is a college student at the Wyss Institute and HMS,
Kramme partnered with co-first author and fellow Church laboratory member Alexandru Plesa, who was experiencing similar disappointments making gene vectors for his task. Kramme, Plesa, and Chatterjee then set to work describing what would be needed to make an end-to-end platform for hereditary screening that would work for all of their tasks, which varied from protein engineering to fertility and aging.
From bits to the bench
To enhance the earliest phase of hereditary research study– determining genes of interest to study– the group produced 2 brand-new algorithms to assist fulfill the requirement for computational tools that can evaluate and draw out details from the progressively big datasets that are being produced through next-generation sequencing (NGS). The very first algorithm takes the basic information about a gene’s expression level and integrates it with details about the state of the cell, in addition to details about which proteins are understood to engage with the gene. The algorithm offers a high rating to genes that are extremely linked to other genes and whose activity is associated with big, cell-level modifications. The 2nd algorithm offers more top-level insight by creating networks to represent the vibrant modifications in gene expression throughout cell-type distinction and after that using midpoint steps, such as Google’s PageRank algorithm, to rank the crucial regulators of the procedure.
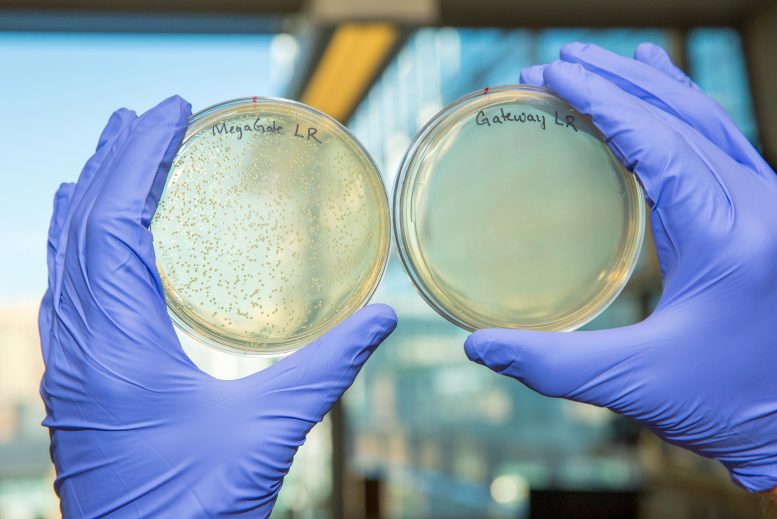
MegaGate, an unique technique for cloning target genes of interest into vectors, is far more effective at producing effective gene-bearing vectors (left) than other existing techniques like Gateway (right). Credit: Wyss Institute at Harvard University
“The computational part of genetic studies is like a Jenga game: if each block in the tower represents a gene, we’re looking for the genes that make up the base of the Jenga tower, the ones that hold the whole thing up. Most algorithms can only tell you which genes are in the same row as each other, but ours allow you to home in on how far up or down the tower they are, so you can quickly identify the ones that have the biggest influence on the cell state in question,” stated Chatterjee.
Once the target genes have actually been determined, the STAMPScreen procedure relocations from the laptop computer to the laboratory, where experiments are carried out to interrupt those genes in cells and see what impact that perturbation has on the cell. The group of scientists methodically examined several gene perturbation tools consisting of complementary DNA (cDNA) and numerous variations of CRISPR in human induced pluripotent stem cells (hiPSCs), the very first recognized head-to-head contrasts carried out completely in this extremely flexible yet tough cell type.
They then produced a brand-new tool that permits CRISPR and cDNA to be utilized within the exact same cell to unlock synergies in between the 2 techniques. For example, CRISPR can be utilized to switch off expression of all isoforms of a gene, and cDNA can be utilized to sequentially reveal each isoform separately, enabling more nuanced hereditary research studies and considerably decreasing background expression of off-target genes.
Scanning library barcodes
The next action in numerous hereditary experiments is creating a screening library for presenting genes into cells and observing their results. Typically, gene pieces are placed into bacterial plasmids (circular pieces of DNA) utilizing techniques that work well for little pieces of DNA, however are troublesome to utilize when placing bigger genes. Many of the existing techniques likewise count on a method called Gateway, which utilizes a procedure called lambda phage recombination and the production of a toxic substance to exterminate any germs that did not get a plasmid with the gene of interest. The contaminant in these plasmids is typically troublesome to deal with in the laboratory, and can be unintentionally suspended when a “barcode” series is contributed to a vector to assist scientists determine which gene-bearing plasmid the vector got.
Kramme and Plesa were dealing with Gateway when they recognized that these issues might be fixed if they got rid of the contaminant and changed it with brief series on the plasmid that would be acknowledged and cut by a kind of enzyme called meganucleases. Meganuclease acknowledgment series do not appear in the genes of any recognized organism, hence guaranteeing that the enzyme will not unintentionally cut the placed gene itself throughout cloning. These acknowledgment series are naturally lost when a plasmid gets a gene of interest, making those plasmids unsusceptible to meganuclease. Any plasmids that do not effectively get the gene of interest, nevertheless, maintain these acknowledgment series and are cut to pieces when a meganuclease is included, leaving just a pure swimming pool of plasmids including the placed gene. The brand-new technique, which the scientists called MegaGate, had a cloning success rate of 99.8% and likewise enabled them to barcode their vectors with ease.
“MegaGate not only solves many of the problems that we kept running into with older cloning methods, it is also compatible with many existing gene libraries like the TFome and hORFeome. You can essentially take Gateway and meganucleases off the shelf, put them together with a library of genes and a library of barcoded destination vectors, and two hours later you have your barcoded genes of interest. We’ve cloned nearly 1,500 genes with it, and have yet to have a failure,” stated Plesa, who is a college student at the Wyss Institute and HMS.
Finally, the scientists showed that their barcoded vectors might be effectively placed into living hiPSCs, and swimming pools of cells might be evaluated utilizing NGS to identify which provided genes were being revealed by the swimming pool. They likewise effectively utilized a range of techniques, consisting of RNA-Seq, TAR-Seq, and Barcode-Seq, to check out both the hereditary barcodes and the whole transcriptomes of hiPSCs, making it possible for scientists to utilize whichever tool they are most knowledgeable about.
The group prepares for that STAMPScreen might show beneficial for a wide array of research studies, consisting of path and gene regulative network research studies, distinction aspect screening, drug and complex path characterizations, and anomaly modeling. STAMPScreen is likewise modular, enabling researchers to incorporate various parts of it into their own workflows.
“There’s a treasure trove of information housed in publicly available genetic datasets, but that information will only be understood if we use the right tools and methods to analyze it. STAMPScreen will help researchers get to eureka moments faster and speed up the pace of innovation in genetic engineering,” stated senior author George Church,Ph D., a Wyss Core Faculty member who is likewise a Professor of Genetics at HMS and Professor of Health Sciences and Technology at Harvard and MIT.
“At the Wyss Institute we aim for impactful ‘moonshot’ solutions to pressing problems, but we know that to get to the moon, we have to first build a rocket. This project is a great example of how our community innovates on-the-fly to enable scientific breakthroughs that will change the world for the better,” stated Wyss Founding Director Don Ingber, M.D.,Ph D., who is likewise the Judah Folkman Professor of Vascular Biology at HMS and the Vascular Biology Program at Boston Children’s Hospital, in addition to Professor of Bioengineering at Harvard John A. Paulson School of Engineering and Applied Sciences.
Reference: “An Integrated Pipeline for Mammalian Genetic Screening” 27 September 2021, Cell Reports Methods
Additional authors of the paper consist of Helen Wang, Bennett Wolf, Merrick Smela, Xiaoge Guo,Ph D., and Richie Kohman,Ph D. from the Wyss Institute and HMS.