The waterborne algae Chlamydomonas reinhardtii. Credit: Image by He et al
Carbon is among the primary foundation for life on Earth. It’s plentiful in our world’s environment, where it’s discovered in the kind of co2. Carbon makes its method into Earthlings’ bodies generally through the procedure of photosynthesis, which includes co2 into sugars that work as elements for crucial biomolecules and sustain the international food cycle. About a 3rd of this procedure internationally is performed by single-celled algae that reside in the oceans (the majority of the rest is done by plants).
The enzyme that carries out the primary step of the response to absorb co2 into sugars is a large protein called Rubisco put together from 8 similar little subunits and 8 similar big subunits organized together symmetrically. All the parts of this assembly, which is called a holoenzyme, operate in performance to carry out Rubisco’s enzymatic task. Rubisco’s rate of activity — and by extension, the rate at which plants and algae can grow — is restricted by its access to co2. Free co2 can be limited in water, so marine algae such as Chlamydomonas reinhardtii in some cases battle to keep Rubisco operating at peak capability. To neutralize this, these algae progressed an unique structure called the pyrenoid to provide focused co2 to Rubisco. The pyrenoid is so crucial that nearly all algae on earth have one. Different types of algae are believed to have actually progressed the structure individually.
“The defining feature of a pyrenoid is the matrix, a giant liquid-like condensate that contains nearly all of the cell’s Rubisco,” discusses Jonikas, an Assistant Professor in the Department of Molecular Biology at Princeton.
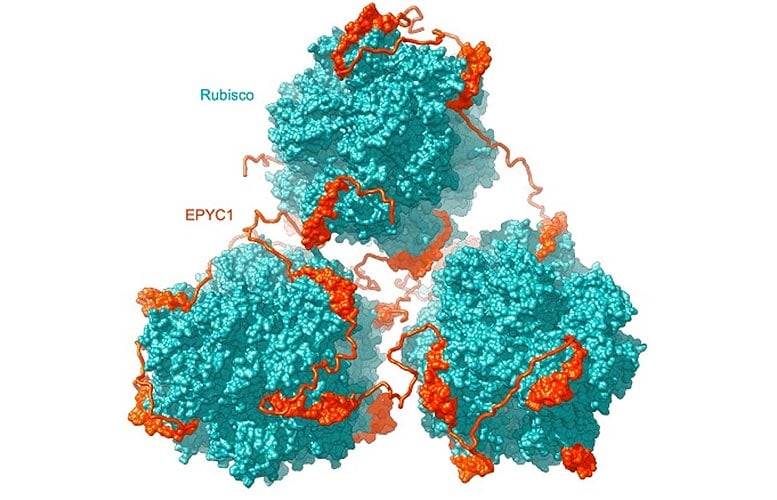
Rubisco (teal) is connected and clustered together by EPYC1 (orange) in the pyrenoid matrix. Subhead: Princeton scientists Shan He, Martin Jonikas, and their associates have actually found how Rubisco holoenzymes put together to form the fluid-like matrix of the algal pyrenoid, an organelle that moderates the incorporation of co2 into sugars. The research study detailing the group’s findings was released November 23, 2020 in the journal Nature Plants. Credit: Image by He et al.
Rubisco is the primary element of the pyrenoid matrix, however not the only one; in 2016, Jonikas’s laboratory found another plentiful protein in the pyrenoid called EPYC1. In their 2016 paper, Jonikas’s group revealed that EPYC1 binds to Rubisco and assists concentrate Rubisco in the pyrenoid. The scientists thought that EPYC1 works like a molecular glue to connect together Rubisco holoenzymes. Postdoc Shan He, together with associates in Jonikas’s laboratory and partners from Germany, Singapore and England, set out to check this theory.
“In the present work, we demonstrate that this is indeed how it works,” states Jonikas, “by showing that EPYC1 has five binding sites for Rubisco, allowing it to ‘link’ together multiple Rubisco holoenzymes.”
EPYC1 is a loosely structured, extended protein, and its 5 Rubisco binding websites are equally dispersed throughout its length. The scientists likewise discovered that Rubisco has 8 EPYC1 binding websites dispersed equally throughout its ball-like surface area. Computer modeling revealed that the loosely structured and versatile EPYC1 protein can make numerous contacts with a single Rubisco holoenzyme or bridge together surrounding ones. In in this manner, EPYC1 drives Rubisco to cluster in the pyrenoid matrix.
Although this provides a rewarding description for how the matrix is put together, it postures something of a dilemma. Other proteins require to be able to gain access to Rubisco to fix it when it breaks down. If the EPYC1-Rubisco network is stiff, it might obstruct these proteins from accessing Rubisco. However, He and associates discovered that EPYC1’s interactions with Rubisco are relatively weak, so although the 2 proteins might form lots of contacts with each other, these contacts are exchanging quickly.
“This allows EPYC1 and Rubisco to flow past each other while staying in a densely packed condensate, allowing other pyrenoid proteins to also access Rubisco,” notes Jonikas. “Our work solves the longstanding mystery of how Rubisco is held together in the pyrenoid matrix”.
Land plants don’t have pyrenoids, and researchers believe that engineering a pyrenoid-like structure into crop plants might enhance their development rates. Understanding how the pyrenoid is put together in algae represents a substantial action towards such efforts.
“He and colleagues provide a very nice molecular study of the protein-protein interactions between the Rubisco small subunit and EPYC1,” states Dr. James Moroney, Professor of Biology at the Louisiana State University department of Biological Sciences, whose laboratory research studies photosynthesis in plants and algae.
“This work is encouraging for researchers trying to introduce pyrenoid-like structures into plants to improve photosynthesis,” he includes.
In a world beleaguered by cravings and illness, we can utilize all the increases we can get.
Reference: “The structural basis of Rubisco phase separation in the pyrenoid” by Shan He, Hui-Ting Chou, Doreen Matthies, Tobias Wunder, Moritz T. Meyer, Nicky Atkinson, Antonio Martinez-Sanchez, Philip D. Jeffrey, Sarah A. Port, Weronika Patena, Guanhua He, Vivian K. Chen, Frederick M. Hughson, Alistair J. McCormick, Oliver Mueller-Cajar, Benjamin D. Engel, Zhiheng Yu and Martin C. Jonikas, 23 November 2020, Nature Plants.
DOI: 10.1038/s41477-020-00811-y
Funding: The work explained here was supported by grants to M.C.J. from the National Science Foundation (nos IOS-1359682 and MCB-1935444), National Institutes of Health (no. DP2-GM-119137), and Simons Foundation and Howard Hughes Medical Institute (no. 55108535); to B.D.E. by Deutsche Forschungsgemeinschaft (EN 1194/1-1 as part of FOR2092); to O.M.-C. by the Ministry of Education (MOE Singapore) Tier 2 (no. MOE2018-T2-2-059); to A.J.M. and N.A. by the UK Biotechnology and Biological Sciences Research Council (no. BB/ S015531/1) and Leverhulme Trust (no. RPG-2017-402); to F.M.H by NIH ( R01GM071574); to S.A. P. by Deutsche Forschungsgemeinschaft fellowship (no. PO2195/1-1); and to V.K.C. by a National Institute of General Medical Sciences of the Institutes of Health (no. T32GM007276) training grant.