Supercomputer simulations at Los Alamos National Laboratory showed that the G type of SARS-CoV-2, the dominant pressure of the infection triggering COVID-19, altered to a conformation that permits it to more quickly connect to host receptors, while likewise being more prone to antibodies than the initial D type. Credit: Los Alamos National Laboratory
Dominant G-form Spike protein ‘puts its head up’ more regularly to lock on to receptors, however that makes it more susceptible to neutralization.
Large-scale supercomputer simulations at the atomic level reveal that the dominant G type version of the COVID-19-triggering infection is more transmittable partially since of its higher capability to easily bind to its target host receptor in the body, compared to other variations. These research study arises from a Los Alamos National Laboratory–led group brighten the system of both infection by the G type and antibody resistance versus it, which might assist in future vaccine advancement.
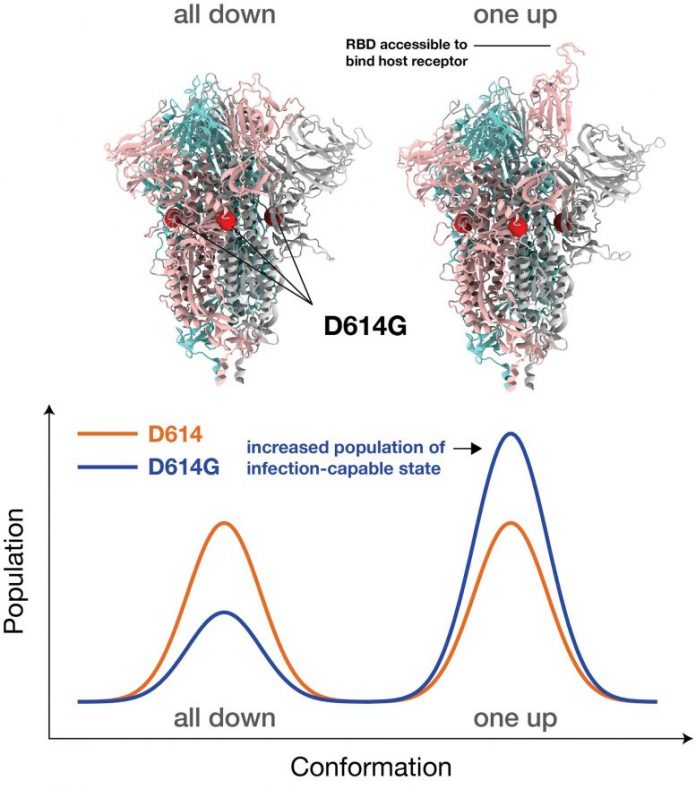
“We found that the interactions among the basic building blocks of the Spike protein become more symmetrical in the G form, and that gives it more opportunities to bind to the receptors in the host — in us,” stated Gnana Gnanakaran, matching author of the paper released just recently in Science Advances. “But at the same time, that means antibodies can more easily neutralize it. In essence, the variant puts its head up to bind to the receptor, which gives antibodies the chance to attack it.”
Researchers understood that the version, likewise referred to as D614G, was more transmittable and might be reduced the effects of by antibodies, however they didn’t understand how. Simulating more than a million specific atoms and needing about 24 million CPU hours of supercomputer time, the brand-new work supplies molecular-level information about the habits of this version’s Spike.
Current vaccines for SARS-CoV-2, the infection that triggers COVID-19, are based upon the initial D614 type of the infection. This brand-new understanding of the G version — the most comprehensive supercomputer simulations of the G type at the atomic level — might suggest it provides a foundation for future vaccines.
The group found the D614G version in early 2020, as the COVID-19 pandemic triggered by the SARS-CoV-2 infection was increase. These findings were released in Cell. Scientists had actually observed an anomaly in the Spike protein. (In all variations, it is the Spike protein that provides the infection its particular corona.) This D614G anomaly, called for the amino acid at position 614 on the SARS-CoV-2 genome that went through an alternative from aspartic acid, dominated internationally within a matter of weeks.
The Spike proteins bind to a particular receptor discovered in a lot of our cells through the Spike’s receptor binding domain, eventually causing infection. That binding needs the receptor binding domain to shift structurally from a closed conformation, which cannot bind, to an open conformation, which can.
The simulations in this brand-new research study show that interactions amongst the foundation of the Spike are more balanced in the brand-new G-form version than those in the initial D-form pressure. That balance results in more viral Spikes outdoors conformation, so it can quicker contaminate an individual.
A group of postdoctoral fellows from Los Alamos — Rachael A. Mansbach (now assistant teacher of Physics at Concordia University), Srirupa Chakraborty, and Kien Nguyen — led the research study by running numerous microsecond-scale simulations of the 2 variations in both conformations of the receptor binding domain to brighten how the Spike protein engages with both the host receptor and with the reducing the effects of antibodies that can assist secure the host from infection. The members of the research study group likewise consisted of Bette Korber of Los Alamos National Laboratory, and David C. Montefiori, of Duke Human Vaccine Institute.
The group thanks Paul Weber, head of Institutional Computing at Los Alamos, for supplying access to the supercomputers at the Laboratory for this research study.
Reference: “The SARS-CoV-2 Spike variant D614G favors an open conformational state” by Rachael A. Mansbach, Srirupa Chakraborty, Kien Nguyen, David C. Montefiori, Bette Korber, S. Gnanakaran, 16 April 2021, Science Advances.
DOI: 10.1126/sciadv.abf3671
Funding: The task was supported by Los Alamos Laboratory Directed Research and Development task 20200706ER, Director’s Postdoctoral fellowship, and the Center of Nonlinear Studies Postdoctoral Program at Los Alamos.