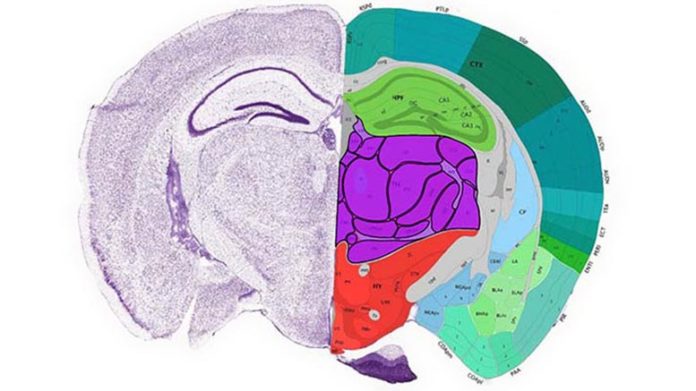
The Mouse Brain Atlas is a multi-year, multi-institutional effort to parse the genomics underlying kind and function of the mouse brain, which acts as a design for associated human research study. Photo credit: Allen BrainInstitute Credit: Allen Brain Institute
The circuits of the human brain include more than 100 billion nerve cells, each connected to lots of other nerve cells through countless synaptic connections, leading to a three-pound organ that is exceptionally more complicated than the amount of its many parts.
In current years, nevertheless, transformative advances in imaging, sequencing and computational innovations have actually opened the possibility of mapping a human brain really at the resolution of its molecular and cellular parts. While that supreme objective stays to be accomplished, scientists have actually gradually advanced with a smaller sized, however no less special, effort: an atlas of the mouse brain.
In an unique concern of Nature, scientists at the University of California San Diego, with associates throughout the nation, explain their development in collection of documents. Two of the documents, in which UC San Diego researchers worked as senior authors, even more fine-tune the company of cells within essential areas of the mouse brain and, more seriously, the company of transcriptomic, epigenomic, and regulative aspects and aspects that offer these brain cells with function and function.
“To truly understand how the brain functions, and from that knowledge develop new drugs and therapies to improve human lives and health, we need to see and quantify brain structure, organization and function down to the level of single cells,” stated Bing Ren, PhD, director of the Center for Epigenomics, teacher of cellular and molecular medication at UC San Diego School of Medicine and member of the Ludwig Institute for Cancer Research at UC San Diego.
“Depth and specificity are essential,” concurred Eran A. Mukamel, PhD, director of the Computational Neural DNA Dynamics Lab and associate teacher in the Department of Cognitive Science at UC SanDiego “We want a comprehensive parts list for the brain, including not just the locations and connections of the neurons, but also the molecular and epigenetic fingerprints that give them their specialized identity.”
Gene regulative aspects
Since 2006, there has actually been a collective, global effort to produce a three-dimensional atlas of the mouse brain, which is approximately the size of a pea and consisted of around 8 to 14 million nerve cells and glial cells. Though the mouse brain is not a mini variation of the human brain, it has actually shown to be an effective design for studying lots of human brain functions, illness, and mental illness, in part since the genes accountable for structure and running both human and rodent organs are 90 percent similar.
In their paper, senior author Ren, associates, and partners at the Center for Epigenomics concentrated on developing an atlas of gene regulative aspects in the mouse cerebrum, the evolutionarily youngest area of the brain that supports top-level sensory understanding, motor control, and cognitive functions.
Recent studies of mouse and human brains have actually exposed that the cerebrum includes numerous neural cell types dispersed in various areas, however the transcriptional regulative programs– the instructions accountable for each cell’s distinct pattern of gene expression, and thus its identity and function– stay unidentified.
Ren’s group penetrated available chromatin– the things of chromosomes– in more than 800,000 private cell nuclei from 45 places in the adult mouse brain, then utilized the information to map the state of 491,818 prospect cis-regulatory DNA aspects in 160 unique cell types. Cis- regulative aspects are areas of non-coding DNA that control transcription (copying a section of DNA into RNA) of surrounding genes.
They discovered that various kinds of nerve cells lie in unique locations of the mouse brain, and the uniqueness of their spatial circulation and function is associated, and most likely driven, by the distinct set of cis-regulatory DNA aspects within each cell type. Indeed, a few of the cell-type-specific aspects recognized by Ren’s group were separately revealed to be enough to drive press reporter gene expression in particular sub-classes of nerve cells in the mouse brain.
Surprisingly, the majority of the mouse brain cis-regulatory aspects mapped by the scientists have homologous or comparable series in the human genome that might serve as regulative aspects, and for that reason might be utilized to annotate gene regulative aspects associated with human brain cell type requirements.
Ren stated the findings offer a structure for extensive analysis of gene regulative programs of the mammalian brain, consisting of human beings, and can help in translating noncoding danger variations that add to different neurological illness and characteristics in human beings.
Transcriptomic and epigenomic aspects
Each cell or population of cells produces a distinct pattern of RNA records– hairs of RNA transcribed from DNA that communicate hereditary guidelines for the proteins that direct and sustain life. It’s approximated that countless chain reactions take place within mammalian cells every second. That intricacy, integrated with growing datasets explaining the functions of genes, fats, proteins, sugars, and other gamers in cell biology, have actually made complex efforts to comprehend how the brain is arranged and functions.
Mukamel and associates combined advanced sequencing strategies to concentrate on the mouse main motor cortex, a brain area basic to motion. They created more than 500,000 transcriptomes and epigenomes– extensive listings of all of the RNA particles and adjustments of DNA that make each mouse brain cell distinct.
Using unique computational and analytical designs, they produced a multimodal atlas of 56 neuronal cell key ins the mouse main motor cortex that thoroughly explains their molecular, genomic, and structural functions.
Mukamel stated the research study revealed that each brain cell has actually a collaborated pattern of gene expression and epigenetic policy that can be acknowledged with high fidelity utilizing various sequencing strategies. Just as a person has particular handwriting, facial functions, singing patterns and personality type, the authors discovered that the RNA and DNA signatures of cell key ins the motor cortex distinguish each cell from its next-door neighbors.
And simply as our human uniqueness adds to the strength and variety of our neighborhoods, stated Mukamel, the distinct patterns of gene expression and policy in brain circuits support an extremely varied network of cells with specialized functions and synergistic functions.
By integrating both epigenomic and transcriptomic information from an extraordinary variety of cells, Mukamel stated the research study shows the capacity of single-cell sequencing innovations to thoroughly map brain cell types– a lesson that will assist in comprehending the more complicated circuits of the human brain.
References:
“An atlas of gene regulatory elements in adult mouse cerebrum” by Yang Eric Li, Sebastian Preissl, Xiaomeng Hou, Ziyang Zhang, Kai Zhang, Yunjiang Qiu, Olivier B. Poirion, Bin Li, Joshua Chiou, Hanqing Liu, Antonio Pinto-Duarte, Naoki Kubo, Xiaoyu Yang, Rongxin Fang, Xinxin Wang, Jee Yun Han, Jacinta Lucero, Yiming Yan, Michael Miller, Samantha Kuan, David Gorkin, Kyle J. Gaulton, Yin Shen, Michael Nunn, Eran A. Mukamel, M. Margarita Behrens, Joseph R. Ecker and Bing Ren, 6 October 2021, Nature
DOI: 10.1038/ s41586-021-03604 -1
“A transcriptomic and epigenomic cell atlas of the mouse primary motor cortex” by Zizhen Yao, Hanqing Liu, Fangming Xie, Stephan Fischer, Ricky S. Adkins, Andrew I. Aldridge, Seth A. Ament, Anna Bartlett, M. Margarita Behrens, Koen Van den Berge, Darren Bertagnolli, Hector Roux de Bézieux, Tommaso Biancalani, A. Sina Booeshaghi, Héctor Corrada Bravo, Tamara Casper, Carlo Colantuoni, Jonathan Crabtree, Heather Creasy, Kirsten Crichton, Megan Crow, Nick Dee, Elizabeth L. Dougherty, Wayne I. Doyle, Sandrine Dudoit, Rongxin Fang, Victor Felix, Olivia Fong, Michelle Giglio, Jeff Goldy, Mike Hawrylycz, Brian R. Herb, Ronna Hertzano, Xiaomeng Hou, Qiwen Hu, Jayaram Kancherla, Matthew Kroll, Kanan Lathia, Yang Eric Li, Jacinta D. Lucero, Chongyuan Luo, Anup Mahurkar, Delissa McMillen, Naeem M. Nadaf, Joseph R. Nery, Thuc Nghi Nguyen, Sheng-Yong Niu, Vasilis Ntranos, Joshua Orvis, Julia K. Osteen, Thanh Pham, Antonio Pinto-Duarte, Olivier Poirion, Sebastian Preissl, Elizabeth Purdom, Christine Rimorin, Davide Risso, Angeline C. Rivkin, Kimberly Smith, Kelly Street, Josef Sulc, Valentine Svensson, Michael Tieu, Amy Torkelson, Herman Tung, Eeshit Dhaval Vaishnav, Charles R. Vanderburg, Cindy van Velthoven, Xinxin Wang, Owen R. White, Z. Josh Huang, Peter V. Kharchenko, Lior Pachter, John Ngai, Aviv Regev, Bosiljka Tasic, Joshua D. Welch, Jesse Gillis, Evan Z. Macosko, Bing Ren, Joseph R. Ecker, Hongkui Zeng and Eran A. Mukamel, 6 October 2021, Nature
DOI: 10.1038/ s41586-021-03500 -8
Funding came, in part, from the National Institutes of Health (grant U19 MH11483), the Howard Hughes Medical Institute, National Institutes of Health BRAIN Initiative (grants U19 MH114830, U19 MH121282, U19 MH114821, R24 MH114788, U24 MH114827, R24 MH114815) National Institute on Deafness and Other Communication Disorders (DC013817), the Hearing Health Foundation and the National Institute of General Medical Sciences (GM114267).